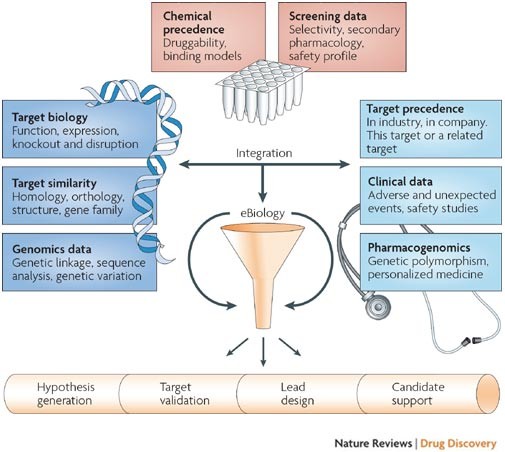
High-throughput electronic biology: mining information for drug discovery
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
KEY POINTS * In the past 10 years, the life sciences have seen a proliferation of electronic data, emerging from systems such as databases, text-mining technologies, high-throughput
techniques and 'omics' platforms (for example, DNA microarray). * In this article, we review some of the recent developments in the field of electronic biology (eBiology), which
uses these resources as a substrate for new drug discovery. As extensive reviews on data sets and tools already exist, we highlight how these resources can be applied directly to solve
bottlenecks in the industry. * A number of approaches to the application of eBiology in drug discovery are identified, ranging from deep 'systems biology' to
'project-specific' analyses and focus on high-throughput techniques. * A set of examples are given, which look at the power of applying multiple resources simultaneously to build
layers of evidence and end with the identification of novel drug targets. In these scenarios, the expert in that disease area is a key partner, without which the exercise is unlikely to
succeed. * Although there are an increasing number of examples of target mining in the literature, there is also a need to consider how one then translates these biological hypotheses into
drug discovery programmes. To this end, we consider the data sources and techniques that can be used to apply a business focus to the results of this mining. * As hypotheses turn into real
programmes, we consider further workflows to support these later stages of discovery, looking at issues such as druggability, selectivity and understanding the action of a compound both _in
vitro_ and _in vivo_. Although these areas are traditionally addressed by computational chemists, there is much to be gained by the use of techniques and resources familiar to eBiologists. *
Last, we discuss the future needs in eBiology and how the area must be primarily led through scientific creativity, rather than technical considerations. ABSTRACT The vast range of _in
silico_ resources that are available in life sciences research hold much promise towards aiding the drug discovery process. To fully realize this opportunity, computational scientists must
consider the practical issues of data integration and identify how best to apply these resources scientifically. In this article we describe _in silico_ approaches that are driven towards
the identification of testable laboratory hypotheses; we also address common challenges in the field. We focus on flexible, high-throughput techniques, which may be initiated independently
of 'wet-lab' experimentation, and which may be applied to multiple disease areas. The utility of these approaches in drug discovery highlights the contribution that _in silico_
techniques can make and emphasizes the need for collaboration between the areas of disease research and computational science. Access through your institution Buy or subscribe This is a
preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per
year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated
during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS HIGH-CONTENT
CRISPR SCREENING Article 10 February 2022 MOVING TARGETS IN DRUG DISCOVERY Article Open access 19 November 2020 DECODING DISEASE: FROM GENOMES TO NETWORKS TO PHENOTYPES Article 02 August
2021 REFERENCES * Searls, D. B. Data integration — connecting the dots. _Nature Biotech._ 8, 844–845 (2003). Google Scholar * Searls, D. B. Data integration: challenges for drug discovery.
_Nature Rev. Drug Discov._ 4, 45–58 (2005). CAS Google Scholar * US Government Accountability Office. _New Drug Development: Science, Business, Regulatory, and Intellectual Property Issues
Cited as Hampering Drug Development Efforts_. _US Government Accountability Office web site_ [online], (2006). * Blagosklonny, M. V. & Pardee, A. B. Conceptual biology: unearthing the
gems. _Nature_ 416, 373 (2002). CAS PubMed Google Scholar * Weeber, M. et al. Generating hypotheses by discovering implicit associations in the literature: a case report of a search for
new potential therapeutic uses for thalidomide. _J. Am. Med. Inform. Assoc._ 10, 252–259 (2003). AN IMPORTANT PAPER THAT HIGHLIGHTS ONE OF THE EBIOLOGY FUNDAMENTALS — HOW NEW HYPOTHESES CAN
BE GENERATED THROUGH MINING TECHNIQUES THAT FOCUS ON THE IDENTIFICATION OF PREVIOUSLY UNKNOWN INDIRECT RELATIONSHIPS BETWEEN ENTITIES. PubMed PubMed Central Google Scholar * Smith U.M. et
al. The transmembrane protein meckelin (MKS3) is mutated in Meckel–Gruber syndrome and the wpk rat. _Nature Genet._ 38, 191–196 (2006). CAS PubMed Google Scholar * Laaksonen R, et al. A
systems biology strategy reveals biological pathways and plasma biomarker candidates for potentially toxic statin-induced changes in muscle. _PLoS ONE_ 1, e97 (2006). PubMed PubMed Central
Google Scholar * Kumar, N. et al. Applying computational modeling to drug discovery and development. _Drug Discov. Today_ 11, 806–811 (2006). CAS PubMed Google Scholar * Cho, C. R. et
al. The application of systems biology to drug discovery. _Curr. Opin. Chem. Biol._ 10, 294–302 (2006). CAS PubMed Google Scholar * Butte, A. & Kohane, I. Creation and implications of
a phenome–genome network. _Nature Biotech._ 24, 55–62 (2006). DESCRIBES HOW LARGE-SCALE COMBINATIONS OF GENE-EXPRESSION ANNOTATION, PHENOTYPE AND ENVIRONMENTAL DATA CAN REVEAL NEW INSIGHTS
INTO DISEASE PROCESSES. CAS Google Scholar * Kim, H. & Dafna, B. Modulation of signalling of sprouty: a developing story _Nature Rev. Mol. Cell Biol._ 5, 441–450 (2004). CAS Google
Scholar * Schoeberl, B. et al. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. _Nature Biotech._ 20, 370–375 (2002).
ILLUSTRATES HOW SYSTEMS APPROACHES CAN TRACK COMPLEX SIGNALLING PATHWAYS IMPORTANT TO CANCER. HIGHLIGHTS THE ESSENTIAL NEED FOR _IN SILICO_ ANALYSIS OF METABOLIC NETWORKS. Google Scholar *
Galperin M. Y. The molecular biology database collection: 2007 update. _Nucleic Acids Res._ 35 (database issue), D3–D4 (2006). PubMed PubMed Central Google Scholar * Stromback, L. et al.
Representing, storing and accessing molecular interaction data: a review of models and tools. _Brief. Bioinformatics_ 7, 331–338 (2006). PubMed Google Scholar * Ashburner, M. et al. Gene
Ontology: tool for the unification of biology. The Gene Ontology Consortium _Nature Genet._ 25, 25–29 (2003). Google Scholar * Torr-Brown, S. Advances in knowledge management for
pharmaceutical research and development. _Curr. Opin. Drug Discov. Devel._ 8, 316–322 (2005). CAS PubMed Google Scholar * Neumann, E. K. & Quan, D. Biodash: a semantic web dashboard
for drug development. _Pac. Symp. Biocomp._ 11, 176–187 (2006). Google Scholar * Blake, J. A. & Bult, C. J. Beyond the data deluge: data integration and bio-ontologies. _J. Biomed.
Inform._ 39, 314–320 (2006). PubMed Google Scholar * Potts S. J., Edwards D. J. & Hoffman R. Challenges of target/compound data integration from disease to chemistry: a case study of
dihydrofolate reductase inhibitors. _Curr. Drug Discov. Technol._ 2, 75–87 (2005). CAS PubMed Google Scholar * Erhardt, R. A., Schneider, R. & Blaschke, C. Status of text-mining
techniques applied to biomedical text. _Drug Discov. Today_ 11, 315–325 (2006). CAS PubMed Google Scholar * Hu, Y. et al. Analysis of genomic and proteomic data using advanced literature
mining. _J. Proteome Res._ 2, 405–412 (2003). DEMONSTRATION OF LITERATURE ANALYSIS ON A GENOME-WIDE SCALE, WHICH HIGHLIGHTS HOW THE USE OF POWERFUL TECHNOLOGIES ACROSS LARGE REPOSITORIES CAN
BE USED TO RAPIDLY IDENTIFY ASSOCIATIONS BETWEEN BIOLOGICAL CONCEPTS. PubMed Google Scholar * Wlodek, D. & Gonzales, M. Decreased energy levels can cause and sustain obesity. _J.
Theor. Biol._ 225, 33–44 (2003). PubMed Google Scholar * Pospisil, P., Iyer, L. K., Adelstein, S. J. & Kassis, A. I. A combined approach to data mining of textual and structured data
to identify cancer-related targets. _BMC Bioinformatics_ 7, 354–365 (2006). PubMed PubMed Central Google Scholar * Stegmann, J. & Grohmann, G. Hypothesis generation guided by co-word
clustering. _Scientometrics_ 56, 111–135 (2003). CAS Google Scholar * Wren, J. D., Bekeredjian, R., Stewart, J. A., Shohet, R. V. & Garner H. R. Knowledge discovery by automated
identification and ranking of implicit relationships. _Bioinformatics_ 20, 389–398 (2004). CAS PubMed Google Scholar * Carley, D. W. Drug repurposing: identify, develop and commercialize
new uses for existing or abandoned drugs. _Part I. Drugs_ 8, 306–309 (2005). PubMed Google Scholar * Marks, D. J. et al. Defective acute inflammation in Crohn's disease: a clinical
investigation. _Lancet_ 367, 668–678 (2006). CAS PubMed Google Scholar * Habashi, J. P. et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome.
_Science_ 312, 117–121 (2006). CAS PubMed PubMed Central Google Scholar * Ozcan, U. et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2
diabetes. _Science_ 313, 1137–1140 (2006). PubMed PubMed Central Google Scholar * Krauthammer, M. et al. Molecular triangulation: bridging linkage and molecular-network information for
identifying candidate genes in Alzheimer's disease. _Proc. Natl Acad. Sci. USA_ 101, 15148–15153 (2004). CAS PubMed Google Scholar * Tiffin, N. et al. Computational disease gene
identification: a concert of methods prioritizes type 2 diabetes and obesity candidate genes. _Nucleic Acids Res._ 34, 3067–3081 (2006). A HIGHLY PERTINENT EXAMPLE OF HOW GENOME MAPPING AND
MULTIPLE LINES OF DATA AND EVIDENCE CAN BE COMBINED WITH DISEASE ASSOCIATION TO IDENTIFY POTENTIAL NOVEL GENE LINKAGES WITH DIABETES, AND THEREBY SUPPORT WIDE-RANGING HYPOTHESIS GENERATION.
CAS PubMed PubMed Central Google Scholar * Owens, D., Grimley, J. & Kirkpatrick, P. Inhaled human insulin. _Nature Rev. Drug Discov._ 5, 371–372 (2006). CAS Google Scholar *
Fitzgerald, G. A. Anticipating change in drug development: the emerging era of translational medicine and therapeutics. _Nature Rev. Drug Discov._ 4, 815–818 (2005). CAS Google Scholar *
Liu J. J. et al. Multiclass cancer classification and biomarker discovery using GA-based algorithms. _Bioinformatics_ 21, 2691–2697 (2005). CAS PubMed Google Scholar * Mittleman, B. B.
Biomarkers for systemic lupus erythematosus: has the right time finally arrived? _Arthritis Res. Ther._ 6, 223–224 (2004). PubMed PubMed Central Google Scholar * Baker, M. In biomarkers
we trust? _Nature Biotech._ 23, 297–304 (2005). CAS Google Scholar * Lindsay, M. Finding new drug targets in the 21st century. _Drug Discov. Today_ 10, 1683–1687 (2005). CAS PubMed
Google Scholar * Kling, J. From hypertension to angina to Viagra. _Modern Drug Discov._ 1, 31–38 (1998). CAS Google Scholar * Mullins, I. M. et al. Data mining and clinical data
repositories: insights from a 667,000 patient data set. _Comput. Biol. Med._ 36, 1351–1377 (2006). PubMed Google Scholar * Reynolds G. P. et al. The 5-HT2C receptor and
antipsychoticinduced weight gain — mechanisms and genetics. _J. Psychopharmacol._ 20, 15–18 (2006). PubMed Google Scholar * Song, J. et al. Development of homogeneous high-affinity agonist
binding assays for 5-HT2 receptor subtypes. _Assay Drug Dev. Technol._ 3, 649–659 (2005). CAS PubMed Google Scholar * Jensen, L. J., Saric, J. & Bork, P. Literature mining for the
biologist: from information retrieval to biological discovery. _Nature Rev. Genet._ 7, 119–129 (2006). CAS PubMed Google Scholar * Swanson, D. R. On the fragmentation of knowledge, the
connection explosion, and assembling other people's ideas. ASIST Award of Merit acceptance speech. _Bulletin ASIST_ 27, 12–14 (2001). Google Scholar * Hopkins, A. L. & Groom, C. R.
The druggable genome. _Nature Rev. Drug Discov._ 1, 727–730 (2002). CAS Google Scholar * Lui, J. & Rost, B. Target space for structural genomics revisited. _Bioinformatics_ 18,
922–933 (2002). Google Scholar * Vinod, P. K., Konkimalla, B. & Chandra, N. _In-silico_ pharmacodynamics: correlation of adverse effects of H2-antihistamines with histamine _N_-methyl
transferase binding potential. _Appl. Bioinformatics_ 5, 141–150 (2006). CAS PubMed Google Scholar * Hajduk, P. J., Huth, J. R. & Fesik, S. W. Druggability indices for protein targets
derived from NMR-based screening data. _J. Med. Chem._ 48, 2518–2525 (2005). CAS PubMed Google Scholar * An, J., Totrov, M. & Abagyan R. Comprehensive identification of
'druggable' protein ligand binding sites. _Genome Inform._ 15, 31–41 (2004). CAS PubMed Google Scholar * Cheng, A. et al. Structure-based maximal affinity model predicts
small-molecule druggability analysis. _Nature Biotech._ 25, 71–75 (2007). Google Scholar * Nair, R. & Rost, B. LOCnet and LOCtarget: sub-cellular localization for structural genomics
targets. _Nucleic Acids Res._ 32, W517–W521 (2004). CAS PubMed PubMed Central Google Scholar * Froloff, N. et al. in _Chemogenomics: Knowledge-based Approaches to Drug Discovery_ (ed.
Jacoby, E.) 175–206 (Imperial College Press, London, 2006). Google Scholar * Birault, V. et al. Bringing kinases into focus: efficient drug design through the use of chemogenomic toolkits.
_Curr. Med. Chem._ 13, 1735–1748 (2006). CAS PubMed Google Scholar * Hug, H. et al. Ontology-based knowledge management of troglitazone-induced hepatotoxicity. _Drug Discov. Today_ 9,
948–953 (2004). PubMed Google Scholar * Frye, S. V. Structure–activity relationship homology (SARAH): a conceptual framework for drug discovery in the genomic era. _Chem. Biol._ 6, R3–R7
(1999). AN EXCELLENT EXAMPLE OF A POWERFUL GENERIC WORKFLOW THAT REUSES EXISTING STRUCTURE–ACTIVITY RELATIONSHIP DATA TO ADDRESS THE IMPORTANT ISSUE OF TARGET SELECTIVITY. CAS PubMed
Google Scholar * Nettles J. H. et al. Bridging chemical and biological space: 'target fishing' using 2D and 3D molecular descriptors. _J. Med. Chem._ 49, 6802–6810 (2006).
DESCRIBES A TARGET FISHING WORKFLOW THAT USES THE WEALTH OF INFORMATION IN LARGE STRUCTURE–ACTIVITY RELATIONSHIP DATABASES TO UNDERSTAND MOLECULAR ACTIVITIES. CAS PubMed Google Scholar *
Schuffenhauer, A. & Jacoby, E. Annotating and mining the ligand-target chemogenomics knowledge space. _Drug Discov. Today: BioSilico_ 2, 190–200 (2004). CAS Google Scholar * Eriksson
T., Bjorkman S., Roth B. & Hoglund P. Intravenous formulations of the enantiomers of thalidomide: pharmacokinetic and initial pharmacodynamic characterization in man. _J. Pharm.
Pharmacol._ 52, 807–817 (2005). Google Scholar * Kalgutkar A. S. & Soglia J. R. Minimising the potential for metabolic activation in drug discovery. _Expert Opin. Drug Metab. Toxicol._
1, 91–142 (2005). CAS PubMed Google Scholar * Spinks, D. & Spinks, G. Serotonin reuptake inhibition: an update on current research strategies. _Curr. Med. Chem._ 9, 799–810 (2002).
CAS PubMed Google Scholar * Sanderson, D. M. & Earnshaw, C. G. Computer prediction of possible toxic action from chemical structure; The DEREK system. _Human Exp. Toxicol._ 10,
261–273 (1991). CAS Google Scholar * Patlewicz, G., Rodford, R. & Walker, J. D. Quantitative structure-activity relationships for predicting mutagenicity and carcinogenicity. _Environ.
Toxicol. Chem._ 22, 1885–1893 (2003). CAS PubMed Google Scholar * Greene N. Computer systems for the prediction of toxicity: an update. _Adv. Drug Deliv. Rev._ 31, 417–431 (2002). Google
Scholar * Mayne, J. T., Ku, W. W. & Kennedy, S. P. Informed toxicity assessment in drug discovery: systems-based toxicology. _Curr. Opin. Drug Discov. Devel._ 9, 75–83 (2006). CAS
PubMed Google Scholar * Niculescu, S. P., Atkinson, A., Hammond G. & Lewis M. Using fragment chemistry data mining and probabilistic neural networks in screening chemicals for acute
toxicity to the fathead minnow. _SAR QSAR Environ. Res._ 15, 293–309 (2004). CAS PubMed Google Scholar * Krejsa, C. M. et al. Predicting ADME properties and side effects: the BioPrint
approach. _Curr. Opin. Drug Discov. Devel._ 6, 470–480 (2003). CAS PubMed Google Scholar * White, A. C., Mueller, R. A., Gallavan, R. H., Aaron, S. & Wilson, A. G. A multiple _in
silico_ program approach for the prediction of mutagenicity from chemical structure. _Mutat. Res._ 5, 77–89 (2003). Google Scholar * Dearden, J. C. _In silico_ prediction of drug toxicity.
_J. Comput. Aided Mol. Des._ 17, 119–127 (2003). CAS PubMed Google Scholar * Snyder, R. D. et al. Assessment of the sensitivity of the computational programs DEREK, TOPKAT, and MCASE in
the prediction of the genotoxicity of pharmaceutical molecules. _Environ. Mol. Mutagen._ 43, 143–158 (2004). CAS PubMed Google Scholar * Fliri, A. F., Loging, W. T., Thadeio, P. &
Volkmann, R. A. Biological spectra analysis: linking biological activity profiles to molecular structure. _Proc. Natl Acad. Sci. USA_ 102, 261–266 (2005). DESCRIBES A NEW APPROACH TO
UNDERSTANDING THE 'PROTEOME INTERACTION POTENTIAL' FOR SMALL MOLECULES; TERMED 'BIOSPECTRA', WHICH ALLOWS FOR THE GROUPING OF COMPOUNDS AND THEIR POTENTIAL PROPERTIES.
CAS PubMed Google Scholar * Fliri, A. F., Loging, W. T., Thadeio, P. & Volkmann, R. A. Analysis of drug-induced effect patterns linking structure and side effects of medicine's.
_Nature Chem. Biol._ 1, 389–397 (2005). CAS Google Scholar * Fliri, A. F., Loging, W. T., Thadeio, P. & Volkmann, R. A. Biospectra analysis: model proteome characterizations for
linking molecular structure and biological response. _J. Med. Chem._ 48, 6918–6925 (2005). CAS PubMed Google Scholar * Weggen, S. et al. A subset of NSAIDs lower amyloidogenic Aβ42
independently of cyclooxygenase activity. _Nature_ 414, 212–216 (2001). CAS PubMed Google Scholar * Lehmann, J. et al. Redesigning drug discovery. _Nature_ 384 (Suppl. 6604), 1–5 (1996).
Google Scholar * Lamb, J. et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. _Science_ 29, 1929–1935 (2006). A RECENT EFFORT
DESIGNED TO MAKE USE OF GENOMICS DATA DIRECTLY IN A DRUG DISCOVERY APPLICATION BY PROVIDING THE ABILITY TO SCREEN COMPOUNDS AGAINST GENOME-WIDE DISEASE SIGNATURES. Google Scholar * Fielden,
M. R. & Kolaja, K. L. The state-of-the-art in predictive toxicogenomics. _Curr. Opin. Drug Discov. Devel._ 9, 84–91 (2006). CAS PubMed Google Scholar * Quackenbush, J. Top-down
standards will not serve systems biology. _Nature_ 440, 24 (2006). CAS PubMed Google Scholar * Southern, E. M. Detection of specific sequences among DNA fragments separated by gel
electrophoresis. _J. Mol. Biol._ 98, 503–517 (1975). CAS PubMed Google Scholar * Kleppe, K., Ohtsuka, E., Kleppe, R., Molineux, I. & Khorana, H. G. Studies on polynucleotides. XCVI.
Repair replications of short synthetic DNA's as catalyzed by DNA polymerases. _J. Mol. Biol._ 56, 341–361 (1971). CAS PubMed Google Scholar * Mullis, K., et al. Specific enzymatic
amplification of DNA _in vitro_: the polymerase chain reaction. _Cold Spring Harb. Symp. Quant. Biol._ 51, 263–273 (1986). CAS PubMed Google Scholar * Kell, D. B. & Oliver, S. G. Here
is the evidence now what is the hypothesis? The complimentary role of inductive and hypothesis-driven science in the post-genomic era. _Bioessays_ 26, 99–109 (2004). Google Scholar * Pao,
M. L. _Concepts of Information Retrieval_ (Libraries Unlimited, Englewood, Colorado, 1989). Google Scholar * Gund, P., Maliski, E. & Brown, F. The evolution of pharmaceutical
informatics as a discipline _Curr. Opin. Drug Discov. Devel._ 9, 301–302 (2006) CAS Google Scholar * Swanson, D. R. Fish oil, Raynaud's syndrome, and undiscovered public knowledge.
_Perspect. Biol. Med._ 30, 7–18 (1986). A FUNDAMENTAL PAPER THAT DESCRIBES HOW EXISTING MEDICAL KNOWLEDGE CAN HARBOUR INDIRECT LINKAGES, WHICH CAN BE EXPLOITED TO IDENTIFY AND GENERATE
NOVEL, TESTABLE HYPOTHESES. CAS PubMed Google Scholar * Nasukawa, T. & Nagano, T. Text analysis and knowledge mining system. _IBM Systems J._ 40, 967–984 (2001). Google Scholar *
Rhodes et al. Mining patents using molecular similarity search. _Pac. Symp. Biocomput._ 12, 304–315 (2007). Google Scholar Download references ACKNOWLEDGEMENTS The authors would like to
thank T. Turi, J. Lanfear, S. Campbell, I. Harrow, J. Keeling and the Pfizer PharmaMatrix team for their valuable insight and support while preparing this article. We also wish to gratefully
acknowledge the input and suggestions from the reviewers. Finally, we wish to acknowledge the contribution of those who develop and maintain a vast landscape of _in silico_ resources and
apologize to those who we have been unable to cite in this article. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Computational Biology Group, Pfizer, Groton, Connecticut, USA William Loging
* Computational Biology Group, Pfizer, Sandwich, Kent, UK Lee Harland * eBiology Group, Pfizer, Sandwich, Kent, UK Bryn Williams-Jones Authors * William Loging View author publications You
can also search for this author inPubMed Google Scholar * Lee Harland View author publications You can also search for this author inPubMed Google Scholar * Bryn Williams-Jones View author
publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to William Loging. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing financial interests. RELATED LINKS RELATED LINKS DATABASES OMIM Alzheimer disease Crohn disease Marfan syndrome Parkinson disease type 2 diabetes FURTHER INFORMATION BioCarta
Biomarkers.org Ensembl Gene Expression Omnibus Gene Ontology Mouse Genome Informatics database _Nature Biotechnology_ Community Consultation Online Mendelian Inheritance in Man Protein Data
Bank Simplified Molecular Input Line Entry Specification GLOSSARY * _In silico_ A term used to describe experiments or experimental results that are held electronically. * Unified medical
language system Controlled compendium of vocabularies across the spectrum of life sciences. * Medical subject heading The US National Library of Medicine's controlled vocabulary used
for indexing articles for Medline. * Single nucleotide polymorphism A specific location in a DNA sequence at which different people can have a different DNA base. Differences in a single
base could change the protein sequence, leading to disease (for example, sickle-cell disease), or have no known consequences. * Translational medicine The testing of novel therapeutic
strategies (in humans) that were developed through basic laboratory experimentation. Observations taken 'from the bedside to the bench' also constitute translational medicine. *
RSS RSS is a family of data formats used to publish frequently updated digital content, such as news feeds and alerts from many web sites on the internet. * Quantitative structure–activity
relationships (QSAR). Mathematical relationships linking chemical structure and pharmacological activity in a quantitative manner for a series of compounds. Methods that can be used in QSAR
include various regression and pattern-recognition techniques. * Ames positivity A biological assay that assesses DNA damage caused by small molecule drugs in bacterial cells. RIGHTS AND
PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Loging, W., Harland, L. & Williams-Jones, B. High-throughput electronic biology: mining information for drug
discovery. _Nat Rev Drug Discov_ 6, 220–230 (2007). https://doi.org/10.1038/nrd2265 Download citation * Issue Date: March 2007 * DOI: https://doi.org/10.1038/nrd2265 SHARE THIS ARTICLE
Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided
by the Springer Nature SharedIt content-sharing initiative