Mutation analysis of cbp and pcaf reveals rare inactivating mutations in cancer cell lines but not in primary tumours
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT In this study we screened the histone acetyltransferases CBP and PCAF for mutations in human epithelial cancer cell lines and primary tumours. We identified two CBP truncations
(both in cell lines), seven PCAF missense variants and four CBP intronic microdeletions. These data suggest that neither gene is commonly inactivated in human epithelial cancers. SIMILAR
CONTENT BEING VIEWED BY OTHERS ONCOHISTONE MUTATIONS ENHANCE CHROMATIN REMODELING AND ALTER CELL FATES Article 01 March 2021 PARALLEL FUNCTIONAL ANNOTATION OF CANCER-ASSOCIATED MISSENSE
MUTATIONS IN HISTONE METHYLTRANSFERASES Article Open access 02 November 2022 METTL3 PROMOTES TUMOUR DEVELOPMENT BY DECREASING APC EXPRESSION MEDIATED BY _APC_ MRNA
_N_6-METHYLADENOSINE-DEPENDENT YTHDF BINDING Article Open access 21 June 2021 MAIN The addition of an acetyl group to specific lysine residues within the N-terminal region of the four core
histone proteins by acetyltransferases, causes the destabilisation of the chromatin structure and enhances access of transcription factors and other DNA-binding components to DNA (Grunstein,
1997). The histone acetyltransferases CBP, P300 and PCAF also acetylate sequence-specific transcription factors such as P53. CBP was originally isolated on the basis of its interaction with
CREB in response to cAMP signalling (Chrivia et al, 1993). P300 was purified as a cellular protein, which binds the adenoviral protein E1A (Eckner et al, 1994). PCAF, P300/CBP-associated
factor, was the first mammalian histone acetyltransferase discovered on the basis of homology to yeast Gcn5p (Yang et al, 1996). The fact that histone acetyltransferases are involved in cell
proliferation and differentiation suggests that they may be involved in cancer. Indeed _P300_ (also known as _EP300_) and _CBP_ are fused to _MLL_ in acute myeloid leukaemia. It is also
known that P300, CBP and PCAF are targeted by viral oncoprotein E1A (Eckner et al, 1994; Yang et al, 1996; Chakravarti et al, 1999). In colorectal and gastric carcinomas two somatic _P300_
missense mutations coupled to deletion of the second allele of the gene were identified (Muruoka et al, 1996). The role of _P300_ as a tumour suppressor gene was later confirmed with the
identification of truncating, insertion and missense mutations in primary tumours and cancer cell lines, associated with inactivation of the second allele (Gayther et al, 2000). In this
study we screened the whole coding sequence and intron–exon boundaries of both _CBP_ and _PCAF_ for somatic mutations in a series of human primary tumours and cancer cell lines. We also
screened a panel of cell lines for truncating _P300_ mutations using Western blotting. MATERIALS AND METHODS SAMPLES The _CBP_ gene was screened in 179 DNA samples isolated from 59 primary
breast tumours, 37 primary ovarian tumours, 20 colorectal tumours, and 63 cancer cell lines. The _PCAF_ gene was screened in 80 cancer cell lines (31 breast, 25 ovarian, 10 pancreatic, 6
SCLC, 5 colorectal, 1 NSCLC, 1 MISC, 1 BCLL) and 20 primary colorectal tumours. In all cases the collection of tumour material was done with Local Research Ethics Committee approval. All
tumours were ‘flash’ frozen immediately following surgery. Cell lines were obtained from ATCC and ECACC cell repository or as a gift from collaborating laboratories. PREPARATION OF DNA AND
RNA Frozen primary tumours were serially sectioned onto slides. Tumour tissue was microdissected and DNA extracted by SDS-proteinase K digestion followed by phenol-chloroform extraction.
Germ-line DNA was prepared from either a matching blood sample or from normal tissue. Cell line DNA was extracted by either proteinase K or DNAzol™ (Gibco BRL). RNA was extracted with
TriZol™ (Gibco BRL). cDNA was synthesized by reverse transcription of RNA using random hexamers and Superscript II (Gibco BRL). DETERMINATION OF THE EXON–INTRON STRUCTURE OF _CBP_ AND _PCAF_
The exon-intron structure of _CBP_ and _PCAF_ were determined from the available cDNA and genomic DNA sequences in Genbank (NCBI). _CBP_ is a 8694 bp cDNA consisting of 32 exons distributed
over 154 Kb of genomic sequence at chromosome band 16p13.3. PCAF is a 2957 bp cDNA consisting of 20 exons spread over 114 Kb of genomic sequence at chromosome band 3p24. POLYMERASE CHAIN
REACTION _CBP_ was amplified from gDNA in 43 fragments and _PCAF_ was amplified from cDNA in 13 fragments of approximately 200–400 bp (oligonucleotide primer sequences are available on
request, [email protected]). _PCAF_ sequence alterations were confirmed subsequently in genomic DNA. Amplification reactions (30 μl) contained 20 mM (NH4)2SO4, 75 mM TrisHCl, pH 9.0 at 25°C,
0.1% (w v−1) Tween, 2.5–3 mM MgCl2, 200 μM dNTP, 10 pmoles of each primer and 2.5 U of Red Hot DNA polymerase (Advanced Biotechnologies). The amplifications were done using a DNA Engine
Tetrad, MJ Research PTC-225 Peltier Thermal Cycler. PROTEIN TRUNCATION TEST _PCAF_ coding sequence was analysed initially by PTT. Cell lines HCT15 and OVCAR8, which showed an altered sized
P300 protein on Western blot were also analysed by PTT. RT–PCR amplification was done in overlapping fragments of approximately 1000–1200 bp in length each, using a 5′ oligo containing the
appropriate sequences (oligonucleotide sequences are available on request). PTT reactions were performed following the manufacturer's protocol (Promega). Alterations found in PTT were
confirmed by sequencing. SSCP/HA (SINGLE STRAND CONFORMATION POLYMORPHISM/HETERODUPLEX ANALYSIS) Formamide loading buffer was added to PCR products. The mix was denatured at 95°C for 10 min
and kept on ice until loading onto 0.8 × MDE (Mutation Detection Enhancement) gel (Flowgen), both with and/or without 10% Glycerol. Gels were run overnight at 120 V and 4°C. WESTERN BLOT
ANALYSIS Western blot analysis was used to screen for _P300_ truncating mutations in a panel of 24 cell lines. We also performed Western blot in cell lines identified to have truncating
_CBP_ mutations. Cell extracts were prepared by direct lysis on cell culture plates (TBS, 0.5% NP-40, 5 mM EDTA, Complete Protease Inhibitor Coctail, Boehringer), then electrophoresed in
pre-cast polyacrylamide Tris-Glycine gels (Novex). The separated proteins were transferred to nitrocellulose membrane (Millipore) and hybridised with the respective primary (CBP A-22 Santa
Cruz, P300 N-15 Santa Cruz) and secondary antibodies (Dako). Detection employed the ECL kit (Amersham). DNA SEQUENCING Purified PCR products were sequenced using ABI PrismR BigDye
terminators and an ABI377 sequencer or ABI3100 genetic analyzer (Applied Biosystems, Foster, CA, USA). All samples with a mutation were re-amplified and re-sequenced. RESULTS AND DISCUSSION
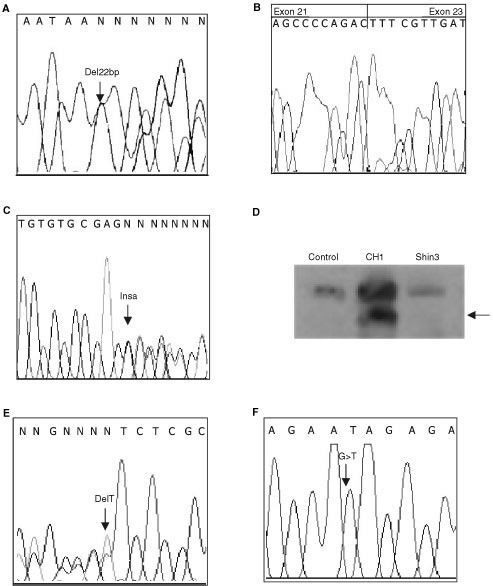
_CBP_ MUTATIONS Two different _CBP_ truncating mutations were identified in the 63 cell lines analysed (Table 1). Shin3, an ovarian cancer cell line, was found to have a heterozygous 22 bp
deletion in intron 21 at position −4 (Figure 1A). This intronic deletion was shown to cause an in-frame deletion of the whole exon 22 at the cDNA level (Figure 1B). In four cancer cell lines
(LK1, LK2, PA1, CH1) an identical heterozygous insertion of an A was found in intron 31 at position −7 (Figure 1C). This insertion was shown to create an alternative splice donor site. This
in turn caused a frame-shift and a premature stop codon at nucleotide 5457 (codon 1795). This heterozygous mutation was confirmed using Western blotting (Figure 1D). The finding of the
identical mutation raised the suspicion of cross contamination between cell lines. HLA typing was performed and the results showed that these cell lines were indeed the same, despite
originating from two different labs (data not shown). We considered these as a single cell line for purposes of mutation frequency analysis and therefore the truncating mutations were
identified in two distinct cell lines out of 60 analysed (3%). No truncating mutations were identified in 116 primary tumours. Small intronic microdeletions in _CBP_ were identified in four
samples (Table 2). A primary colorectal cancer had a heterozygous deletion of T at position −2 in intron 18. This tumour had no molecular phenotype suggestive of microsatellite instability
(MSI). Two cancer cell lines with MSI, OVIP and HCT15, had an identical microdeletion. These intronic microdeletions, which were very close to the splice donor site, had no apparent effect
on mRNA splicing as tested by amplification of cDNA with primers flanking exon19. A breast cancer cell line, MT3, was found to have a deletion of T in intron 14 at position −82, with no
apparent effect on splicing as tested by RT–PCR. This cell line is also MSI+. Uncommon _CBP_ single nucleotide polymorphisms were also detected (Table 3). Although we have characterised two
truncating _CBP_ mutations in cancer cell lines, the absence of inactivating mutations in the primary tumours analysed prevents us from unequivocally establishing the role of _CBP_ in human
primary cancers. Nevertheless the uncommon mutations identified, together with the increased tumour incidence in Rubinstein-Taybi syndrome (Petrij et al, 1995) and the tumorigenic phenotype
in CBP mice (Kung et al, 2000) provide circumstantial evidence for a possible role of _CBP_ as a tumour suppressor gene. We can only speculate on the significance of the intronic
microdeletions seen, but we note that we have previously identified similar intronic microdeletions in other genes in MSI cell lines and primary tumours (data not shown). MISSENSE PCAF
SEQUENCE ALTERATIONS No truncating mutations were identified in the _PCAF_ gene. We used cDNA to screen for truncating mutations and therefore nonsense mediated RNA decay (Maquat, 1995)
could have contributed to a lower sensitivity of the mutation screen. Missense sequence alterations in PCAF were identified in 1 out of 20 primary tumours (5%) and 5 out of 80 cell lines
(6%). In a colorectal cancer case the missense variant was a C to A transversion at nucleotide 2595 in exon 17, resulting in a proline to threonine substitution at codon 713. The same
sequence alteration was also found in the germ-line DNA of the same individual (Table 2). The functional significance of this single nucleotide polymorphism was not tested but this residue
is conserved in mouse and human GCN5, suggesting it might be important for protein function. Sequence analysis in DNA extracted from laser capture microdissected normal and tumour tissue
samples confirmed the heterozygous mutation in both tumour and germ-line DNA, and therefore the mutation is not associated with somatic allelic deletion. A missense alteration at nucleotide
1615 (N386S) was found in four cell lines (Ovmana, Hela, L23, MC4000/ Matu). The mouse homologue of this residue is serine, implying that this alteration is a polymorphism. An ovarian cancer
cell line, OVI-P, had a C to T transition at nucleotide 2415 resulting in Arg653Trp substitution. This arginine residue is conserved in human GCN5, and therefore this substitution could
impair protein function. In addition to these missense sequence alterations a single nucleotide deletion in the 3′ untranslated region of _PCAF_ was found in a colon cancer cell line, SW48.
Three silent _PCAF_ polymorphisms were also identified (Table 3). The missense sequence alteration at nucleotide 1615 has been previously reported and considered a polymorphism (Nishimori et
al, 2000). TRUNCATING _P300_ MUTATIONS We have previously shown that truncating mutations resulted in the production of stable protein, detectable by Western blot. Using this approach we
studied a panel of 24 cell lines and in two (8.3%) truncated protein was detected (Table 1). OVCAR8 had two bands on the Western blot, one of normal size and one from a truncated protein,
suggesting a heterozygous mutation. Sequencing confirmed a heterozygous frameshift deletion (6387delT) resulting in truncation of the protein at codon 1733 (Figure 1E). HCT15 expressed only
a truncated protein and sequencing showed a homozygous transversion, 4239 G → T, generating a stop codon (Figure 1F). This same mutation was previously found in DLD1 as a heterozygous
alteration. Since it is known that DLD1 and HCT15 were derived from the same patient (Chen et al, 1995), one can interpret this result as indicating that the two cell lines were derived from
different areas of the cancer or that the genetic progression represents an event occuring during _in vitro_ culture. The finding of truncating P300 mutations was not a surprise, since we
have previously shown that P300 mutations occur in a small percentage of human epithelial malignancies and others have also identified truncating mutations (Oshima et al, 2001). We have now
analysed a total of 222 cancer samples and truncating mutations have been detected in 6 out of 107 (5.6%) cell lines and 2 out of 115 (1.7%) primary tumours. In conclusion, this report
revealed inactivating CBP mutations in cancer cell lines but not in primary tumours. It also identified a few missense alterations in PCAF and intronic sequence variants in CBP. These data
are insufficient to establish a role for either gene in epithelial cancers. CHANGE HISTORY * _ 16 NOVEMBER 2011 This paper was modified 12 months after initial publication to switch to
Creative Commons licence terms, as noted at publication _ REFERENCES * Chakravarti D, Ogryzko V, Kao H-Y, Nash A, Chen H, Nakatani Y, Evans RM (1999) A viral mechanism for inhibition of p300
and PCAF acetyltransferase activity. _Cell_ 96: 393–403 Article CAS Google Scholar * Chen TR, Dorotinsky CS, McGuire LJ, Macy ML, Hay RJ (1995) DLD-1 and HCT-15 cell lines derived
separately from colorectal carcinomas have totally different chromosome changes but the same genetic origin. _Cancer Genet Cytogenet_ 81: 103–108 Article CAS Google Scholar * Chrivia JC,
Kwok RPS, Lamb N, Hagiwara M, Montminy MR, Goodman RH (1993) Phosphorylated CREB binds specifically to the nuclear protein CBP. _Nature_ 365: 855–859 Article CAS Google Scholar * Eckner
R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingstone DM (1994) Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a
protein with properties of a transcriptional adaptor. _Genes Dev_ 8: 869–884 Article CAS Google Scholar * Gayther SA, Batley S, Linger L, Bannister A, Thorpe K, Chin S, Daigo Y, Russell
P, Wilson A, Sowter H, Delhanty J, Ponder B, Kouzarides T, Caldas C (2000) Mutations truncating the P300 acetylase in human cancers. _Nat Genet_ 24: 300–303 Article CAS Google Scholar *
Grunstein M (1997) Histone aceylation in chromatin structure and transciption. _Nature_ 389: 349–352 Article CAS Google Scholar * Kung AL, Rebel VI, Bronson RT, Cheng LE, Sieff CA,
Livingston DM, Yao TP (2000) Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. _Genes Dev_ 14(3): 272–277 CAS PubMed PubMed Central Google Scholar *
Maquat LE (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. _RNA_ 1: 453–465 CAS PubMed PubMed Central Google Scholar * Muruoka M,
Konishi M, Kikucchi-Yanoshita R, Tanaka K, Shitara N, Chong J-M, Iwama T, Miyaki M (1996) p300 gene alterations in colorectal and gastric carcinomas. _Oncogene_ 12: 1565–1569 Google Scholar
* Nishimori H, Nishikawa R, Fujimaki T, Nakagomi T, Matsutani M, Su Huang H-J, Cavenee WK (2000) Analysis of the p300/CBP-Associated Factor (_PCAF_) gene in astrocytic tumors. _J
Neuro-Oncology_ 46: 17–22 Article CAS Google Scholar * Oshima T, Suganuma T, Ikeda M (2001) A novel mutation lacking the bromodomain of the transcriptional coactivator p300 in the SiHa
cervical carcinoma cell line. _Biochem Biophys Res Commun_ 281: 569–575 Article Google Scholar * Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RCM, Masuno M, Tommerup N, Ommen GJB
(1995) Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. _Nature_ 376: 348–351 Article CAS Google Scholar * Yang X-J, Ogryzko VV, Nishikawa J, Howard
BH, Nakatani Y (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. _Nature_ 382: 319–324 Article CAS Google Scholar Download references ACKNOWLEDGEMENTS
This work was supported by a EU grant and by Cancer Research UK. AUTHOR INFORMATION Author notes * A Försti Present address: Department of Biosciences at Novum, Karolinska Institute, 14157,
Huddinge, Sweden AUTHORS AND AFFILIATIONS * Department of Oncology, Cancer Genomics Program, University of Cambridge, Hutchison/MRC Research Centre, Cambridge, CB2 2XZ, UK H Özdağ, S J
Batley, A Försti, N G Iyer, Y Daigo, B A J Ponder & C Caldas * Wellcome/CRC Institute and Department of Pathology, University of Cambridge, Tennis Court Road, Cambridge, CB2 1QR, UK J
Boutell & T Kouzarides * University of Cambridge Pathology Department, Molecular Histopathology, Addenbrooke's Hospital, Box 235, Level 3, Hills Road, Cambridge, CB2 2QQ, UK M J
Arends Authors * H Özdağ View author publications You can also search for this author inPubMed Google Scholar * S J Batley View author publications You can also search for this author
inPubMed Google Scholar * A Försti View author publications You can also search for this author inPubMed Google Scholar * N G Iyer View author publications You can also search for this
author inPubMed Google Scholar * Y Daigo View author publications You can also search for this author inPubMed Google Scholar * J Boutell View author publications You can also search for
this author inPubMed Google Scholar * M J Arends View author publications You can also search for this author inPubMed Google Scholar * B A J Ponder View author publications You can also
search for this author inPubMed Google Scholar * T Kouzarides View author publications You can also search for this author inPubMed Google Scholar * C Caldas View author publications You can
also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to C Caldas. RIGHTS AND PERMISSIONS From twelve months after its original publication, this work is
licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Özdağ, H., Batley, S., Försti, A. _et al._ Mutation analysis of CBP and PCAF reveals rare inactivating mutations in cancer cell
lines but not in primary tumours. _Br J Cancer_ 87, 1162–1165 (2002). https://doi.org/10.1038/sj.bjc.6600554 Download citation * Received: 19 April 2002 * Revised: 04 July 2002 * Accepted:
31 July 2002 * Published: 29 October 2002 * Issue Date: 04 November 2002 * DOI: https://doi.org/10.1038/sj.bjc.6600554 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative KEYWORDS * P300 * CBP * PCAF * mutations * epithelial cancers