No association between ctnnbl1 and episodic memory performance
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Polymorphisms in the gene encoding catenin-β-like 1 (_CTNNBL1_) were recently reported to be associated with verbal episodic memory performance—in particular, delayed verbal free
recall assessed between 5 and 30 min after encoding—in a genome-wide association study on healthy young adults. To further examine the genetic effects of _CTNNBL1_, we tested for association
between 455 single-nucleotide polymorphisms (SNPs) in or near _CTNNBL1_ and 14 measures of episodic memory performance from three different tasks in 1743 individuals. Probands were part of
a population-based study of mentally healthy adult men and women, who were between 20 and 70 years old and were recruited as participants for the Berlin Aging Study II. Associations were
assessed using linear regression analysis. Despite having sufficient power to detect the previously reported effect sizes, we found no evidence for statistically significant associations
between the tested _CTNNBL1_ SNPs and any of the 14 measures of episodic memory. The previously reported effects of genetic polymorphisms in _CTNNBL1_ on episodic memory performance do not
generalize to the broad range of tasks assessed in our cohort. If not altogether spurious, the effects may be limited to a very narrow phenotypic domain (that is, verbal delayed free recall
between 5 and 30 min). More studies are needed to further clarify the role of _CTNNBL1_ in human memory. SIMILAR CONTENT BEING VIEWED BY OTHERS GENOME-WIDE META-ANALYSES REVEAL NOVEL LOCI
FOR VERBAL SHORT-TERM MEMORY AND LEARNING Article Open access 16 August 2022 A GENOME-WIDE ASSOCIATION STUDY OF THE LONGITUDINAL COURSE OF EXECUTIVE FUNCTIONS Article Open access 10 July
2021 THE CIRCULATING PROTEOME AND BRAIN HEALTH: MENDELIAN RANDOMISATION AND CROSS-SECTIONAL ANALYSES Article Open access 18 May 2024 INTRODUCTION Papassotiropoulos _et al._1 recently
reported that variants in the gene _CTNNBL1_ (encoding catenin-β-like 1, located on chromosome 20 q11.23-q12) are associated with verbal episodic memory performance. The authors reached this
conclusion based on the data from a genome-wide association study on 1073 healthy young adults from Switzerland; they found that the minor allele of single-nucleotide polymorphism (SNP)
rs16986890 was significantly (_P_=7E−08) associated with better verbal episodic memory performance. This result was corroborated in a second and independent cohort of young adults from
Serbia (_n_=524, _P_=0.003), although a different, nontypical experimental paradigm was used to assess episodic memory performance in that data set. In addition to these genetic association
results, the authors also provided evidence from both gene expression data and functional magnetic resonance imaging experiments to support the notion that rs16986890 in _CTNNBL1_ may
account for genotype-dependent differences in memory-related brain functions. Although the results reported by Papassotiropoulos _et al._1 were consistent for young and highly educated
adults, no information was provided as to whether or not they can be generalized to adults with a wider range of ages and more diverse educational backgrounds. Moreover, their findings were
based mainly on relatively simple verbal recall tasks. Because the concept of episodic memory has a number of other aspects beyond verbal recall, a more comprehensive assessment of this
general phenotype is needed for a better understanding of the potential role(s) of genetic determinants in episodic memory. The present study represents the first independent attempt to
replicate the role of rs16986890 in human episodic memory performance following the initial report. In addition, we substantially extend the analyses of Papassotiropoulos _et al._1 by
investigating the potential effects of 454 other SNPs in or near _CTNNBL1_, and by testing a broad range of verbal and nonverbal episodic memory tasks in both young and old adults.
Assessments were performed in a large and independent genome-wide association study sample from Germany (_n_=1743), collected as part of the Berlin Aging Study II. Despite having sufficient
power to replicate findings of the original report, we found no evidence for a genetic link between _CTNNBL1_ and episodic memory performance in our cohort. MATERIALS AND METHODS
PARTICIPANTS Our sample was recruited as participants in the Berlin Aging Study II,2 a multidisciplinary project aiming to identify and characterize genetic, psychological, medical and
socioeconomic factors relevant to human aging in residents of Berlin, Germany. The sample currently comprises 1946 genetically unrelated, mentally healthy individuals, all of whom were of
self-reported Caucasian ancestry. The study was approved by the local Institutional Review Board, and all participants gave signed informed consent before participation. Among the 1743
probands available for analysis in our study, 425 were young adults aged between 20–30 years and the remaining 1318 were old adults aged between 60–70 years. ASSESSMENT OF EPISODIC MEMORY
PERFORMANCE To assess the role of _CTNNBL1_ in human memory, we used 14 quantitative measures of episodic memory derived from: (a) the forward and backward serial recall paradigms;3,4 (b)
associative memory tasks that assessed item memory as well as item–pair recognition;5 and (c) an image recognition test at retention intervals of 2.5 and 1 h.6 A detailed description of the
tasks and traits analyzed can be found in Supplementary Table 1. For a description on how multicollinearity among traits is dealt with, see description of the statistical analysis procedures
below. FORWARD AND BACKWARD SERIAL RECALL TASK In this task, participants were presented with six different lists of 12 words each. After the presentation of the last item in each list,
participants were asked to recall each word at its correct position. Word lists 1–3 were recalled in forward order (from the first word to the last word of the list), whereas word lists 4–6
were recalled in backward order. Recall was self-paced. For each list, responses were scored using a strict serial recall criterion: an accurate response required that both the word and its
serial position were correct. ITEM AND PAIR ASSOCIATIVE EPISODIC MEMORY TASK The item and pair associative episodic memory task (henceforth labeled as the ‘item–pair’ task) had four
conditions that were tested sequentially in one session. During an initial study phase, participants were visually presented with pairs of unrelated words and were instructed to study each
pair under two conditions: either as two single words (item instruction) or as a pair of words (pair instruction). The study phase of each condition contained 30 pairs of semantically
unrelated words. In the test phrase, subjects in one condition (item recognition) were asked whether they had seen the presented word during the study phase. Half of the words were old
(target items), and the other half were new (distractor items). In the second condition (associative recognition), subjects had to decide whether a presented pair of words had been presented
during the study phase. Half of the presented word pairs were old (target pairs), and the others were formed by recombining words in the previously studied lists (rearranged distractor
pairs). Taken together, by crossing over the two instruction conditions with the two test conditions, the task resulted in four conditions that assessed item memory (item–item and pair–item
tests) and associative memory (pair–pair and item–pair tests). Recognition memory performance was measured as hits minus false alarms to minimize the effects of potential individual
differences in response bias. IMAGE RECOGNITION MEMORY WITH RETENTION Performance in the image recognition memory task was assessed at two retention intervals: 2.5 h and 1 week. At the
beginning of the first session, participants were presented with 48 complex, colored images of scenes of neutral emotional valence; all were derived from the International Affective Picture
System.7 The images were encoded incidentally: during the study phase, participants were required to determine whether the scene was ‘indoor’ or ‘outdoor’—there were 24 scenes in each
category—without explicit requirement of memorization. During retrieval, participants viewed each image for 3 sec and were asked to determine whether each scene had been presented (‘old’) or
not (‘new’) during encoding. In each retrieval test, 24 unique old scenes and 24 unique new scenes (lures) were presented. Taking response bias into account, memory performance was measured
as hits minus false alarms. GENOTYPING, SNP IMPUTATION AND QUALITY CONTROL PROCEDURES DNA of each participant was extracted from whole blood using standard procedures, and it was then
subject to microarray-based SNP genotyping using the Affymetrix ‘Genome-Wide Human SNP Array 6.0’. Before imputation, SNPs violating Hardy–Weinberg equilibrium at _P_⩽1E−06 and those with a
call rate <98%—two commonly used quality control filters—were excluded. This resulted in 829 344 autosomal SNPs in 1946 participants. Among these individuals, 214 were excluded from
subsequent analysis. Each of them had at least one of the following conditions: (a) missing information on age or years of education; (b) <95% call rate; (c) evidence for sample
duplication, relatedness or contamination; (d) inconsistency between recorded and genotypic sex; (e) excessive heterozygosity; and (f) population outlier, which was determined by the
EIGENSOFT program;8 specifically, because all participants were of self-reported Caucasian descents, we excluded ethnic outliers using the Eigenstrat function in EIGENSOFT with iterative
outlier removal. After the above filtering steps, principal components (PCs) were computed again for the 1743 remaining samples. On the basis of the examination of the scree plot, the first
four PCs were retained and used as covariates for the subsequent association analysis, to adjust for potential residual population stratification. Genome-wide imputation of unobserved
genotypes was carried out on the ‘cleaned’ data set using the IMPUTE v2.3.2 software,9, 10 on the basis of the precompiled ‘1000 Genomes Phase I Integrated Variant Set’ reference panels from
the IMPUTE website (March 2012 release). As suggested by Southam _et al_,11 we also applied post-imputation quality control filtering, including only SNPs with IMPUTE- information
thresholds ⩾0.8 and minor allele frequencies at or above 5%. After this post-imputation filtering, a total of 455 high-quality SNPs, 71 genotyped and 384 imputed, within a ±50 kb window
surrounding the _CTNNBL1_ locus (that is, between start bp 36 272 434 and end bp 36 550 520; hg19 human reference genome assembly) were retained for subsequent statistical analyses.
ASSOCIATION ANALYSES The phenotypes we used for evaluating participants’ episodic memory performance are functionally related and statistically correlated. The 14 phenotypes are correlated
with an average correlation coefficient (r) of 0.46. One strategy to analyze these data is to test each SNP against each of the 14 phenotypes individually. Although this approach is
straightforward, it is limited by not incorporating potentially useful information from the structure of multiple (and partially correlated) phenotypes. To address this issue, we also used a
second approach that can test several correlated phenotypes simultaneously. As suggested by a recent review,12 we first applied principal component analysis (PCA) to condense information in
the phenotypes by extracting a small number of orthogonal variables (that is, the PCs) that were weighted linear combinations of the original phenotypes. The extracted variables, which were
the first three PCs from PCA, were then used for association analyses in place of the original phenotypes. As expected, the three components were correlated to the three different episodic
memory tasks, and altogether they could explain 80% of the phenotypic variance. Association analyses were carried out using the episodic memory measures (that is, each trait individually and
the PCA variables) as quantitative traits in an additive linear model, adjusted for age, gender, years of education, as well as the four PCs to account for potential population
stratification. All analyses were performed separately in the ‘old’ and ‘young’ strata, to avoid stratification problems. Association tests were performed using SNPTEST v.1.3,13 which can
account for the uncertainty of imputed genotype calls via missing data likelihood tests. POWER CALCULATIONS The power of our study was assessed in the young and old subgroups separately and
was based on the reported effect sizes from the original study.1 Monte Carlo simulations were performed with 1000 runs for empirical power calculations. For the one-trait-at-a-time approach,
we used the SimpleM software14, 15 to account for the correlations among the 14 test items and the correlations among SNPs due to linkage disequilibrium.16 The results of this analysis,
that is, the effective number of independent tests, were then used for Bonferroni-correction to account for multiple testing.16 The estimated effective number of independent tests was 11 and
60 across the 14 phenotypes (or traits) and 455 SNPs, respectively. Hence, the experiment-wide corrected alpha level was set to 4.55E−03 (that is, 0.05/11), for testing the association
between at least one of the traits and the previously reported significant SNP rs16986890; and it was 7.58E−05 (that is, 0.05/(11 × 60)) for testing the association between at least one of
the traits and any SNP in the _CTNNBL1_ gene region. Our power to detect an association between at least one of the traits and rs16986890 at the originally reported effect size was between
93% and 100% for the ‘young’ stratum and 100% for the ‘old’ stratum (see Table 1). When we extended our search to the whole _CTNNBL1_ gene region, the power to detect association between at
least one of the traits and any of the 455 SNPs in the _CTNNBL1_ gene region was between 61% and 97% for the ‘young’ stratum and 100% for the ‘old’ stratum (see Table 2). With the PCA
approach, our power to detect association between at least one of the three PCs (that is, the extracted variables from PCA of the 14 traits) and rs16986890 was between 90% and 100% for the
‘young’ stratum and 100% for the ‘old’ stratum; and it was between 56% and 96% for the ‘young’ stratum and 100% for the ‘old’ stratum when testing possible associations between at least one
of the PCs and any SNP in the _CTNNBL1_ gene region. RESULTS After quality control and adjusting for potential population stratification, there was only minimal evidence of _P_-value
inflation: _λ_ ranged from 1.00–1.04 across the 14 traits tested in the ‘young’ and ‘old’ strata. The minor allele frequency of rs16986890 was 0.056 and 0.058 for the ‘young’ and the ‘old’
in our German sample, respectively, which are consistent with data (that is, 0.058) reported by the 1000 Genomes Project for European (CEU) samples.18 As seen in Table 1, there was no
evidence that SNP rs16986890, which elicited the strongest signal in the original report,1 was associated with any of the 14 memory measures after correcting for multiple comparisons.
Adjusted results approached experiment-wide significance (corresponding to a nominal _P_-value of 4.55E−03, see Materials and Methods) for two memory measures (‘delay_1_week’ (_P_=0.0671)
and ‘PC3’ (0.0639)) in the ‘old’ stratum. However, the directions of these effects were opposite to what was reported in the original study, suggesting that worse, instead of better, memory
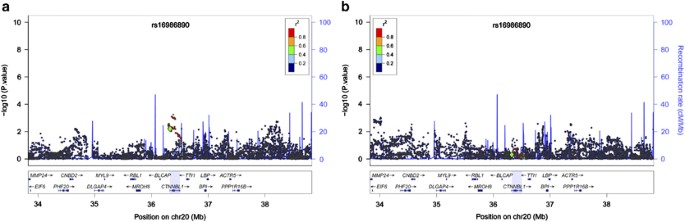
performance was related to the minor allele of rs16986890. Moreover, as seen in the local chromosome region views in Figure 1 and the Supplementary Figures, this ‘signal’ was
indistinguishable from noise. Finally, there was no evidence that any of the other 454 SNPs was significantly associated with the memory measures (all adjusted _P_-values >0.05
(corresponding to a nominal _P_-value of 7.58E−05, see Materials and Methods)). Results obtained using the PCA approach were very similar to those obtained with the one-trait-at-a-time
analyses. Reanalysis of all comparisons without adjusting for years of education—which might themselves have a weak genetic component19 did not change the results appreciably (data not
shown). Summaries of the full results can be found in Table 1, Table 2, Figure 1 and the Supplementary Figures. DISCUSSION In this study, we comprehensively investigated the potential
effects of common genetic variants in or near _CTNNBL1_ on a broad range of verbal and nonverbal episodic memory tasks for both young and old adults. Assessments were performed in over 1700
individuals recruited as part of the Berlin Aging Study II. In contrast to the recently reported findings by Papassotiropoulos _et al._,1 our independent data provide no support for the
notion that SNPs in the _CTNNBL1_ gene region, including rs16986890, exert any significant effects on episodic memory performance. This conclusion holds true regardless of whether we
considered the various tested cognitive traits individually or in combined analyses. Likewise, we saw no evidence of association with respect to age. To the best of our knowledge, this is
the first independent replication attempt since the publication of the original report. On the basis of our data, it remains highly doubtful that genetic variants in _CTNNBL1_ are genuinely
involved in mechanisms controlling episodic memory in humans. The reason for the observed discrepancy between our results and those from the original paper remain elusive. Although we did
not apply the same memory tests (that is, verbal delayed free recall between around 5 and 30 min) as in Papassotiropoulos _et al._,1 our assessments cover a wide range of related tasks
making it unlikely that we have missed a strong general effect of _CTNNBL1_ on episodic memory. Most of the tests applied to our participants can be considered more challenging than the free
recall tests used in the original study. For instance, the serial recall tasks place more demand on associative memory than free recall, which relies mainly on item memory.3,17 Likewise,
the delayed image recognition test used here6 involves a much longer retention interval than 30 min as applied in the original study. Therefore, we cannot rule out the possibility that a
potential effect of SNP rs16986890 (or other SNPs in the _CTNNBL1_ region) is limited to a very narrow phenotypic domain, that is, verbal delayed free recall between around 5 and 30 min. In
summary, our study provides considerable negative evidence against the notion that genetic variants in _CTNNBL1_ are associated with episodic memory performance. More independent assessments
with sufficiently large sample sizes are needed to further clarify the potential role, if any, of this gene in human memory. REFERENCES * Papassotiropoulos A, Stefanova E, Vogler C,
Gschwind L, Ackermann S, Spalek K _et al_. A genome-wide survey and functional brain imaging study identify CTNNBL1 as a memory-related gene. _Mol Psychiatry_ 2011; 18: 255–263. Article
PubMed PubMed Central Google Scholar * Bertram L, Böckenhoff A, Demuth I, Düzel S, Eckardt R, Li S _et al_. Cohort profile: the Berlin Aging Study II (BASE-II). _Int J Epidemiol_ 2014;
43: 703–712. Article PubMed Google Scholar * Lewandowsky S, Murdock B Jr . Memory for serial order. _Psychol Rev_ 1989; 96: 25. Article Google Scholar * Li S, Papenberg G, Nagel I,
Preuschhof C, Schröder J, Nietfeld W _et al_. Aging magnifies the effects of dopamine transporter and D2 receptor genes on backward serial memory. _Neurobiol Aging_ 2013; 34: 358–1. Article
PubMed Google Scholar * Preuschhof C, Heekeren H, Li S, Sander T, Lindenberger U, Bäckman L . KIBRA and CLSTN2 polymorphisms exert interactive effects on human episodic memory.
_Neuropsychologia_ 2010; 48: 402–408. Article PubMed Google Scholar * Papenberg G, Bäckman L, Chicherio C, Nagel I, Heekeren H, Lindenberger U _et al_. Higher intraindividual variability
is associated with more forgetting and dedifferentiated memory functions in old age. _Neuropsychologia_ 2011; 49: 1879–1888. Article PubMed Google Scholar * Lang P Bradley M Cuthbert B .
_International Affective Picture System (IAPS)_, 1st edn. Gainesville, FL, USA: NIMH, Center for the Study of Emotion & Attention, 2005. Google Scholar * Price A, Patterson N, Plenge R,
Weinblatt M, Shadick N, Reich D . Principal components analysis corrects for stratification in genome-wide association studies. _Nat Genet_ 2006; 38: 904–909. Article CAS PubMed Google
Scholar * Marchini J, Howie B . Genotype imputation for genome-wide association studies. _Nat Rev Genet_ 2010; 11: 499–511. Article CAS PubMed Google Scholar * Howie B, Donnelly P,
Marchini J . A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. _PLoS Genet_ 2009; 5: 1000529. Article Google Scholar * Southam
L, Panoutsopoulou K, Rayner N, Chapman K, Durrant C, Ferreira T _et al_. The effect of genome-wide association scan quality control on imputation outcome for common variants. _Eur J Hum
Genet_ 2011; 19: 610–614. Article CAS PubMed PubMed Central Google Scholar * Ott J, Wang J . Multiple phenotypes in genome-wide genetic mapping studies. _Protein Cell_ 2011; 2: 519–522.
Article PubMed PubMed Central Google Scholar * Marchini J, Howie B, Myers S, McVean G, Donnelly P . A new multipoint method for genome-wide association studies by imputation of
genotypes. _Nat Genet_ 2007; 39: 906–913. Article CAS PubMed Google Scholar * Gao X . Multiple testing corrections for imputed SNPs. _Genet Epidemiol_ 2011; 35: 154–158. Article PubMed
PubMed Central Google Scholar * Cheverud J . A simple correction for multiple comparisons in interval mapping genome scans. _Heredity_ 2001; 87: 52–58. Article CAS PubMed Google
Scholar * Nyholt D . A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. _Am J Hum Genet_ 2004; 74: 765–769. Article CAS
PubMed PubMed Central Google Scholar * Li S, Chicherio C, Nyberg L, von Oertzen T, Nagel I, Papenberg G _et al_. Ebbinghaus revisited: influences of the BDNF Val66Met polymorphism on
backward serial recall are modulated by human aging. _J Cogn Neurosci_ 2010; 22: 2164–2173. Article PubMed Google Scholar * 1000 Genomes Project Consortium, Abecasis GR, Altshuler D,
Auton A, Brooks LD, Durbin RM _et al_. A map of human genome variation from population-scale sequencing. _Nature_ 2010; 467: 1061–1073. Article Google Scholar * Rietveld C, Medland S,
Derringer J, Yang J, Esko T, Martin N _et al_. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. _Science_ 2013; 340: 1467–1471. Article CAS
PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This work is supported by the German Federal Ministry of Education and Research (BMBF (grants #16SV5536K,
#16SV5537, #16SV5538 and #16SV5837; previously #01UW0808)). Another source of funding is the Max Planck Institute for Human Development, Berlin, Germany. AUTHOR INFORMATION AUTHORS AND
AFFILIATIONS * Center for Lifespan Psychology, Max Planck Institute for Human Development, Berlin, Germany T Liu, S-C Li, G Papenberg & U Lindenberger * Department of Vertebrate
Genomics, Max Planck Institute for Molecular Genetics, Berlin, Germany T Liu, J Schröder, J T Roehr, W Nietfeld & L Bertram * Department of Psychology, Lifespan Developmental
Neuroscience, TU Dresden, Dresden, Germany, S-C Li * Aging Research Center, Karolinska Institute, Stockholm, Sweden G Papenberg * Charité Universitätsmedizin, Berlin, Germany J Schröder *
School of Public Health, Faculty of Medicine, Imperial College London, London, UK L Bertram Authors * T Liu View author publications You can also search for this author inPubMed Google
Scholar * S-C Li View author publications You can also search for this author inPubMed Google Scholar * G Papenberg View author publications You can also search for this author inPubMed
Google Scholar * J Schröder View author publications You can also search for this author inPubMed Google Scholar * J T Roehr View author publications You can also search for this author
inPubMed Google Scholar * W Nietfeld View author publications You can also search for this author inPubMed Google Scholar * U Lindenberger View author publications You can also search for
this author inPubMed Google Scholar * L Bertram View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to T Liu or L
Bertram. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary Information accompanies the paper on the Translational
Psychiatry website SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (DOC 10014 KB) RIGHTS AND PERMISSIONS This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs
3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the
material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit
http://creativecommons.org/licenses/by-nc-nd/3.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Liu, T., Li, SC., Papenberg, G. _et al._ No association between _CTNNBL1_ and
episodic memory performance. _Transl Psychiatry_ 4, e454 (2014). https://doi.org/10.1038/tp.2014.93 Download citation * Received: 28 January 2014 * Revised: 01 May 2014 * Accepted: 21 May
2014 * Published: 30 September 2014 * Issue Date: September 2014 * DOI: https://doi.org/10.1038/tp.2014.93 SHARE THIS ARTICLE Anyone you share the following link with will be able to read
this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative