Genome-wide methylation analysis identifies specific epigenetic marks in severely obese children
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Obesity is a heterogeneous disease with many different subtypes. Epigenetics could contribute to these differences. The aim of this study was to investigate genome-wide DNA
methylation searching for methylation marks associated with obesity in children and adolescents. We studied DNA methylation profiles in whole blood cells from 40 obese children and controls
using Illumina Infinium HumanMethylation450 BeadChips. After correction for cell heterogeneity and multiple tests, we found that compared to lean controls, 31 CpGs are differentially
methylated in obese patients. A greatest proportion of these CpGs is hypermethylated in obesity and located in CpG shores regions. We next focused on severely obese children and identified
151 differentially methylated CpGs among which 10 with a difference in methylation greater than 10%. The top pathways enriched among the identified CpGs included the “IRS1 target genes” and
several pathways in cancer diseases. This study represents the first effort to search for differences in methylation in obesity and severe obesity, which may help understanding these
different forms of obesity and their complications. SIMILAR CONTENT BEING VIEWED BY OTHERS INTEGRATIVE GENOMIC ANALYSES IN ADIPOCYTES IMPLICATE DNA METHYLATION IN HUMAN OBESITY AND DIABETES
Article Open access 15 May 2023 DNA METHYLATION AND GENE EXPRESSION ANALYSIS IN ADIPOSE TISSUE TO IDENTIFY NEW LOCI ASSOCIATED WITH T2D DEVELOPMENT IN OBESITY Article Open access 19 December
2022 DNA METHYLATION AND ADIPOSITY PHENOTYPES: AN EPIGENOME-WIDE ASSOCIATION STUDY AMONG ADULTS IN THE STRONG HEART STUDY Article 29 July 2020 INTRODUCTION Data from U.S. population surveys
demonstrate a significant increase in obesity prevalence among children age 2–19 years old, from 5.5% in 1976–19801 to 16.9% in 2007–20101,2, with obesity defined as body mass index (BMI) ≥
95th percentile using the Centers for Disease Control and Prevention (CDC)3. Severe obesity is the most rapidly growing paediatric obesity subgroup, and recent estimates suggest that this
disease afflicts up to 6% of all children and adolescents in the United States4. Compared to youth with BMI in the obese range, those with severe obesity have higher rates of immediate and
long-term metabolic and cardiovascular comorbidities5. It stands to reason that youth with obesity and severe obesity may also differ in aetiological factors and consequences, including
epigenetic. There is growing evidence that DNA methylation might contribute to obesity. Indeed, candidate gene methylation studies in animal models and humans have demonstrated methylation
changes in promoters of various genes that are implicated in obesity, appetite control and/or metabolism, insulin signaling, immunity, growth and circadian clock regulation6,7,8,9. For
example, the methylation percentage of _insulin-like growth factor 2 (IGF2_) promoter was higher in overweight infants than in lean infants8. The methylation of _peroxisomal proliferator
activated receptor-γ-co-activator-1α_ promoter in children blood predicts adiposity at adolescence independently of sex, age, pubertal timing and activity6. To identify novel genes and
pathways related to obesity and obesity-induced complications, epigenome-wide association studies (EWAS) are needed. Two previous studies using the HumanMethylation27 BeadChip with 27,000
CpGs, primarily targeting gene promoters and CpG islands (CGIs), examined blood leukocytes of obese and lean adolescents10,11. Wang _et al_. discovered two obesity-associated inflammatory
genes _Ubiquitin Associated And SH3 Domain Containing A_ and _Tripartite Motif Containing 3 (UBASH3A_ and _TRIM3_), and Almen _et al_. identified 20 CpGs differentially methylated between
groups. A larger study looking at the impact of BMI on DNA methylation in different tissues using the HumanMethylation 450 BeadChip by Dick _et al_. found five probes correlated with BMI,
three of which were in the intron of _HIF3A (Hypoxia Inducible Factor 3, Alpha Subunit_)12. In children, two recent papers also showed that specific DNA methylation profiles in blood differ
between lean and obese subjects13,14. Using the NimbleGen Human DNA Methylation 385 K Promoter Plus CpG Island Microarray, Ding _et al_. found 575 demethylated CGIs and 277 hypermethylated
CGIs in obese children. Finally, Huang _et al_. identified 129 differentially methylated CpGs associated with 80 unique genes. None of the obesity-associated CpGs was common to all studies,
which may be due to differences between the study populations, diverse genetic backgrounds, or heterogeneous metabolic phenotypes between different BMI categories. Given the paucity of
research on the different BMI categories, the purpose of this study were twofold: (i) investigate DNA methylation marks in all obese children compare to lean controls; (ii) identify the
differentially methylated CpGs associated with severe obesity in childhood. We hypothesized that patients with severe obesity could help at identifying epigenetic changes, as extreme
phenotypes improve genetic association studies. RESULTS IDENTIFICATION OF DIFFERENTIALLY METHYLATED CPGS ASSOCIATED WITH CHILDHOOD OBESITY The genome-wide methylation analysis was conducted
in 20 obese children (BMI Z-score > 2.5,) and 17 controls (Table 1). Three controls were excluded due to bad quality DNA and arrays. Because DNA methylation varies by cell type and could
bias EWAS results conducted in blood samples, we estimated the cell type compositions in each sample using minfi, and found that cellular composition was not similar between cases and
control subjects (Supplemental Table 1). To correct for these differences, we used next the Houseman’s correction algorithm15. After correction for cellular heterogeneity and multiple
testing, thirty-one CpGs were differentially methylated (FDR ≤ 0.05) between obese children and control subjects, 10 were hypermethylated, and 21 were hypomethylated in obese compared to
lean children (Table 2). The largest difference was observed for cg26834418 located in the promoter region (TSS1500) of the _CHORDC1_ gene (+13% in control subjects compare to obese
children). This gene also showed two other probes differentially methylated in obese compare to lean children. Because of the relatively small numbers in the study and small differences in
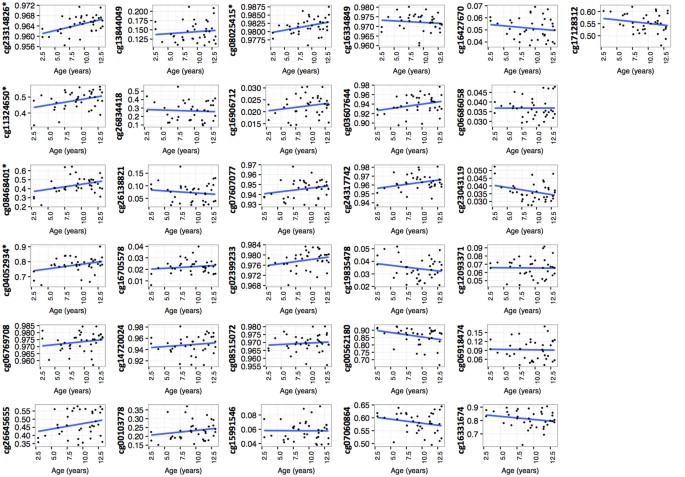
methylation levels for most of the significant CpG sites, we analysed the confounding variables age and sex. We found that the methylation of 5/31 and 1/31 of the identified CpG sites was
correlated with respectively age or sex (Figs 1 and 2). The distribution of the 31 CpGs showed that DNA methylation variation was distributed over the CpG island shores, and that although
CGI are enriched on the array (31% of Illumina probes are in CGI); only 16% of our obesity–associated CpGs were located in CGI, compared to 38% in shores (22% in S-Shore and 16% in N-shore)
(p = 0.016, Pearson’s Chi-squared test) (Fig. 3A). The genomic distribution of the 31 CpGs in comparison to all the probes located on the 450 K BeadChip array with respect to gene structure
is shown in Fig. 3B. We also found an enrichment of differentially methylated CpGs outside promoter and gene body (p = 2.10−4, Pearson’s Chi-squared test) (Fig. 3B). Since obesity is an
extremely heterogeneous disease, we then chose to focus only on the extremely obese children (BMI z-score ≥ 3.5). IDENTIFICATION OF DIFFERENTIALLY METHYLATED CPGS ASSOCIATED WITH SEVERE
CHILDHOOD OBESITY We analysed next DNA methylation marks only in the 11 severely obese children of the group. We found 151 differentially methylated CpGs (q.value ≤ 0.05), 69 were
hypermethylated, and 82 were hypomethylated in severely obese patients compared to lean controls (Supplemental Table 2). Of these 151 differentially methylated CpGs, ten had a greater than
10% difference in methylation between the case and control groups. The most significant difference was observed for a cg27590049 located in the _LMX1A (LIM Homeobox Transcription Factor 1,
Alpha)_16; and the largest difference was observed for a cg07944420 located on the gene body of _ACSF3 (Acyl-CoA Synthetase Family Member 3)_ that showed a decreased by 17% of methylation
level in control subjects compared to obese children. Thirteen of the 151 CpGs were common to our first analysis comparing obese children all together, 18 seems specific to moderately obese
children and 138 to severely obese children (Fig. 3C). Next, we performed a gene set enrichment analysis (GSEA) to explore the potential of shared biologically relevant pathways among the
obesity-associated methylation events. Seven pathways showed a significant enrichment including “IRS1 Target Genes” and different cancer traits (Table 3). The genomic distribution of these
151 differentially methylated CpGs in relation to CpG density (CGIs, shores, shelves, and open sea) was not clearly different from the whole array CpG distribution and there was no
significant enrichment within specific gene regions (data not shown). DISCUSSION In this study we aimed to identify obesity related methylation marks in peripheral blood leukocytes using a
genome wide approach in youth obese children. The primary finding of this study was that most of the epigenetic marks are different in moderate and severe obesity. We identified respectively
18 and 138 differentially methylated CpGs between moderate or severe obese children and lean controls. As observed for genetic association studies, sampling individuals with extreme
phenotypes can enrich the presence of epigenetic variations and can therefore lead to an increase in detection of these differences. Moreover, most of the differentially methylated CpGs was
found within open seas or intergenic regions, consistently with previous findings showing that DNA methylation may be more dynamic outside CGIs. Compare to previous studies10,11,12,13,17,18,
we replicated the association between DNA methylation level and obesity at 10 gene loci (_ANKRD11: ankyrin repeat domain 11, AVPI1: arginine vasopressin-induced 1, CDK19: cyclin-dependent
kinase 19, FOXK2: forkhead box K2, HDAC4: histone deacetylase 4, IFT140: intraflagellar transport 140, KIAA0753, LTBP1: latent transforming growth factor beta binding protein 1, MYOM2:
myomesin 2, TBC1D8: TBC1 domain family, member 8_). Among these genes, _HDAC4_ is an interesting candidate since recently, Abu-Farha _et al_. showed a reduced expression of _HDAC_ mRNA and
protein in human obese subjects both in blood cells and adipose tissue19. This is consistent with clinical data in humans that associated the haploinsufficiency of HDAC4 with obesity20. In
our study, we found a _HDAC_ methylation level higher in obese children than in controls. Numerous genes found in our severe obese analysis were also associated with cancer. Many
epidemiological and clinical studies have demonstrated that early obesity is an established risk factor for many cancers in later life21. Cross talk between macrophages, adipocytes, and
epithelial cells occurs _via_ obesity-associated hormones, growth factor signalling, inflammation, vascular integrity processes, microenvironmental perturbations, and other mediators, which
could enhance the cancer risk and/or progression22. Thus, the methylation changes in childhood obesity could increase the risks for later cancer susceptibility. Most of the identified genes
are not expressed or do not have a relevant function in blood cells; whether these epigenetic marks in blood may reflect or correlate with methylation in more relevant tissues is not known.
However, several studies showed that DNA methylation measured in whole blood is a marker for less accessible tissues that are directly involved in disease. For instance, Murphy _et al_. have
shown for example that methylation across the H19 DMRs (Differentially Methylated Regions) was similar across several tissues from divers embryonic origins23. Likewise, in non-imprinted
loci, Talens _et al_. also found that DNA methylation levels did not differ in blood and buccal cells, from mesodermal and ectodermal embryonic tissues, respectively24. The recent work of
Huang _et al_.25 identified 1,285 discordant and 1,961 concordant genes for methylation between blood and adipose tissue; the discordant genes are enriched in biological functions related to
immune response, leukocyte activation or differentiation, and blood coagulation. Moreover, epigenetic marks associated with type-2 diabetes26 or adiposity7 have also been identified in
peripheral tissues. There were several limitations to this study. The first limitation was that we used DNA from whole blood. To correct our methylation data for this weakness, we used a
Houseman correction algorithm27. It must also be noted that adjusting for cell composition makes impossible the process of replication and validation of the identified CpGs by
pyrosequencing. Replication could also be accomplished if there exists an independent replication population; in this case the models could be re-applied. We tried to replicate our findings
by running the available datasets for common obesity (GEO DataSets: GSE25301, GSE43975, GSE44763, GSE73103) under the RefFreeEwas procedure, but we failed to find any associated CpGs after
correction for cell heterogeneity, even those previously identified by the authors without cell heterogeneity correction. The second limitation is that we cannot conclude about the existence
of these epigenetic marks before the establishment of the obesity. While this lack of interpretability is inherent to the design of the cross-sectional case control study, the finding of
methylation marks associated with the early stages of severe obesity in young patients may be of pathogenic relevance to certain features or complications of this disease. Indeed, the marks
that have been found here could be used in a longitudinal study of the young patients in order to gain both biomarker and mechanistic insights. The longitudinal sampling of cells from
adolescence to adulthood should further allow which of these epigenetic changes follow the development of the long term overt phenotype of severe obesity and its complications. Our major
strength was that all studied participants were children, aged from 3 to 13 years old, less subject to cofounding factors like medication or comorbidities, very common in adult obese
patients. In conclusion, the identification of methylation changes in specific genes will provide important targets for further study into the underlying mechanisms and the therapeutic
potential for childhood obesity. METHODS STUDY PARTICIPANTS Twenty obese children (5 to 13 years old) and equal numbers of control children (3 to 13 years old) were included in the study.
The BMI for obese children (10 male) and lean children (11 male) was 26.0 ± 4.8 kg/m2 and 16.7 ± 2.2 kg/m2, respectively (Table 1). Age and gender-specific BMI Z-scores were determined by
using the growth charts form the World Health Organization with a mean BMI Z-score of 3.6 ± 0.8 for obese children and 0.2 ± 1.1 for controls. Patients with monogenic or syndromic forms of
obesity were excluded. To limit the risk of population stratification, all recruited children are of Caucasian ancestry assessed by family history and grandparents’ birthplace. All methods
were carried out in accordance with relevant guidelines and regulations. Patients and controls were included in the study according to the French bioethics law with families being carefully
informed and having signed a detailed informed consent. All protocols were agreed by French ethic boards (CODECOH DC-2013-1977, CPP C0-13-004, CCTIRS n°14-116bis, CNIL n°91 4228). INFINIUM
HUMANMETHYLATION450 BEADCHIP ARRAY DNA was extracted from whole blood cells of 20 case and 20 control subjects using Gentra DNA extraction kit (Qiagen). Genomic DNA (1 ug) from each of the
40 subjects was bisulphite-converted using Zymo EZ DNA Methylation-Gold kit (ZymoResearch) and the DNA was analysed using the Infinium HumanMethylation450 platform (Illumina, Inc.) by The
Genotyping National Center (CNG, CEA, Evry, France). INFINIUM HUMANMETHYLATION450 BEADCHIP ARRAY DATA PROCESSING DNA methylation status of case and control subjects was established using
Illumina Infinium HumanMethylation450 BeadChips that cover 485,764 cytosine positions of the human genome. Preprocessing and normalization involved steps of probe filtering, color bias
correction, background subtraction and subset quantile normalization as previously described28. After these intra-sample normalization procedures, β-values were calculated. To avoid batch
effect, all samples were processed together. Obese subjects were compared to control subjects, using t-tests. ASSESSMENT OF CELL COMPOSITION DIFFERENCES The R package minfi29 was used to
estimate the fraction of CD8T-, CD4T-, NK- and B-cells, monocytes, and granulocytes in our 40 samples. This package allows for estimating cell fractions in Illumina 450 K methylation data
from whole blood, based on the methylation data published for flow-sorted cells30, and algorithms27. CELL HETEROGENEITY CORRECTION FOR THE METHYLATION DATA ANALYSIS To correct our
methylation data analysis for cell heterogeneity between samples, we used R package RefFreeEwas15. This package allows for conducting EWAS while deconvoluting DNA methylation arising as
mixtures of cell types. This method is similar to surrogate variable analysis, except that it makes additional use of a biological mixture assumption. ADDITIONAL INFORMATION HOW TO CITE THIS
ARTICLE: Fradin, D. _et al_. Genome-Wide Methylation Analysis Identifies Specific Epigenetic Marks In Severely Obese Children. _Sci. Rep._ 7, 46311; doi: 10.1038/srep46311 (2017).
PUBLISHER'S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. REFERENCES * Flegal, K. M., Carroll, M. D.,
Ogden, C. L. & Curtin, L. R. Prevalence and trends in obesity among US adults, 1999–2008. _JAMA_ 303, 235–241, doi: 10.1001/jama.2009.2014 (2010). Article CAS PubMed Google Scholar *
Ogden, C. L., Carroll, M. D., Kit, B. K. & Flegal, K. M. Prevalence of obesity and trends in body mass index among US children and adolescents, 1999–2010. _JAMA_ 307, 483–490, doi:
10.1001/jama.2012.40 (2012). Article PubMed PubMed Central Google Scholar * Ogden, C. L. & Flegal, K. M. Changes in terminology for childhood overweight and obesity. _Natl Health
Stat Report_ 1–5 (2010). * Skinner, A. C. & Skelton, J. A. Prevalence and trends in obesity and severe obesity among children in the United States, 1999–2012. _JAMA Pediatr_ 168,
561–566, doi: 10.1001/jamapediatrics.2014.21 (2014). Article PubMed Google Scholar * Kelly, A. S. et al. Severe obesity in children and adolescents: identification, associated health
risks, and treatment approaches: a scientific statement from the American Heart Association. _Circulation_ 128, 1689–1712, doi: 10.1161/CIR.0b013e3182a5cfb3 (2013). Article PubMed Google
Scholar * Clarke-Harris, R. et al. PGC1alpha promoter methylation in blood at 5-7 years predicts adiposity from 9 to 14 years (EarlyBird 50). _Diabetes_ 63, 2528–2537, doi:
10.2337/db13-0671 (2014). Article PubMed Google Scholar * Drummond, E. M. & Gibney, E. R. Epigenetic regulation in obesity. _Curr Opin Clin Nutr Metab Care_ 16, 392–397, doi:
10.1097/MCO.0b013e3283620f45 (2013). Article CAS PubMed Google Scholar * Perkins, E. et al. Insulin-like growth factor 2/H19 methylation at birth and risk of overweight and obesity in
children. _J Pediatr_ 161, 31–39, doi: 10.1016/j.jpeds.2012.01.015 (2012). Article CAS PubMed PubMed Central Google Scholar * van Dijk, S. J. et al. Epigenetics and human obesity. _Int
J Obes (Lond)_ 39, 85–97, doi: 10.1038/ijo.2014.34 (2015). Article CAS Google Scholar * Wang, X. et al. Obesity related methylation changes in DNA of peripheral blood leukocytes. _BMC
Med_ 8, 87, doi: 1741-7015-8-87 (2010). Article PubMed Central PubMed Google Scholar * Almen, M. S. et al. Genome wide analysis reveals association of a FTO gene variant with epigenetic
changes. _Genomics_ 99, 132–137, doi: S0888-7543(11)00281-3 (2012). Article CAS PubMed Google Scholar * Dick, K. J. et al. DNA methylation and body-mass index: a genome-wide analysis.
_Lancet_ 383, 1990–1998, doi: 10.1016/S0140-6736(13)62674-4 (2014). Article CAS PubMed Google Scholar * Ding, X. et al. Genome-wide screen of DNA methylation identifies novel markers in
childhood obesity. _Gene_ 566, 74–83, doi: 10.1016/j.gene.2015.04.032 (2015). Article CAS PubMed Google Scholar * Huang, R. C. et al. Genome-wide methylation analysis identifies
differentially methylated CpG loci associated with severe obesity in childhood. _Epigenetics_ 10, 995–1005, doi: 10.1080/15592294.2015.1080411 (2015). Article CAS PubMed PubMed Central
Google Scholar * Houseman, E. A., Molitor, J. & Marsit, C. J. Reference-free cell mixture adjustments in analysis of DNA methylation data. _Bioinformatics_ 30, 1431–1439, doi:
10.1093/bioinformatics/btu029 (2014). Article CAS PubMed PubMed Central Google Scholar * German, M. S. et al. Localization of the genes encoding two transcription factors, LMX1 and
CDX3, regulating insulin gene expression to human chromosomes 1 and 13. _Genomics_ 24, 403–404, doi: 10.1006/geno.1994.1639 (1994). Article CAS PubMed Google Scholar * Almen, M. S. et
al. Genome-wide analysis reveals DNA methylation markers that vary with both age and obesity. _Gene_ 548, 61–67, doi: 10.1016/j.gene.2014.07.009 (2014). Article CAS PubMed Google Scholar
* Ollikainen, M. et al. Genome-wide blood DNA methylation alterations at regulatory elements and heterochromatic regions in monozygotic twins discordant for obesity and liver fat. _Clin
Epigenetics_ 7, 39, doi: 10.1186/s13148-015-0073-5 (2015). Article CAS PubMed PubMed Central Google Scholar * Abu-Farha, M. et al. Proteomics analysis of human obesity reveals the
epigenetic factor HDAC4 as a potential target for obesity. _PLoS One_ 8, e75342, doi: 10.1371/journal.pone.0075342 (2013). Article ADS CAS PubMed PubMed Central Google Scholar *
Williams, S. R. et al. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. _Am J Hum
Genet_ 87, 219–228, doi: 10.1016/j.ajhg.2010.07.011 (2010). Article CAS PubMed PubMed Central Google Scholar * Han, X. et al. Body mass index at early adulthood, subsequent weight
change and cancer incidence and mortality. _Int J Cancer_ 135, 2900–2909, doi: 10.1002/ijc.28930 (2014). Article CAS PubMed PubMed Central Google Scholar * Hursting, S. D. Obesity,
energy balance, and cancer: a mechanistic perspective. _Cancer Treat Res_ 159, 21–33, doi: 10.1007/978-3-642-38007-5_2 (2014). Article CAS PubMed Google Scholar * Murphy, S. K., Huang,
Z. & Hoyo, C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. _PLoS One_ 7, e40924, doi: 10.1371/journal.pone.0040924 (2012).
Article ADS CAS PubMed PubMed Central Google Scholar * Talens, R. P. et al. Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology.
_FASEB J_ 24, 3135–3144, doi: fj.09-150490 (2010). Article CAS PubMed Google Scholar * Huang, Y. T. et al. Epigenome-wide profiling of DNA methylation in paired samples of adipose tissue
and blood. _Epigenetics_ 11, 227–236, doi: 10.1080/15592294.2016.1146853 (2016). Article PubMed PubMed Central Google Scholar * Toperoff, G. et al. Genome-wide survey reveals
predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. _Hum Mol Genet_ doi: ddr472 (2011). * Houseman, E. A. et al. DNA methylation arrays as surrogate
measures of cell mixture distribution. _BMC Bioinformatics_ 13, 86, doi: 10.1186/1471-2105-13-86 (2012). Article PubMed PubMed Central Google Scholar * Touleimat, N. & Tost, J.
Complete pipeline for Infinium((R)) Human Methylation 450 K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. _Epigenomics_ 4, 325–341,
doi: 10.2217/epi.12.21 (2012). Article CAS PubMed Google Scholar * Aryee, M. J. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA
methylation microarrays. _Bioinformatics_ 30, 1363–1369, doi: 10.1093/bioinformatics/btu049 (2014). Article CAS PubMed PubMed Central Google Scholar * Reinius, L. E. et al. Differential
DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. _PLoS One_ 7, e41361, doi: 10.1371/journal.pone.0041361 (2012). Article
ADS CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS Our thanks go to the study participants of the Epichild/Obgen study and staff for excellent technical
assistance. This work was supported by an ANR (Agence Nationale de la Recherche) grant: DSS-2012 Epichild and a University Paris Sud (Paris 11) grant: “Attractivité 2015”. AUTHOR INFORMATION
Author notes * Delphine Fradin Present address: Present address: Inserm U1232, CRCINA, Nantes, France., AUTHORS AND AFFILIATIONS * INSERM U1169, Bicêtre Hospital, Paris Sud University, Le
Kremlin-Bicêtre, France Delphine Fradin, Marie-Pierre Belot, Fanny Lachaux & Pierre Bougnères * Pierre et Marie Curie University, INSERM, Paris, U707, France Pierre-Yves Boëlle *
Laboratory for Epigenetics and Environment, Centre National de Génotypage, CEA - Institut de Génomique, Evry, France Jorg Tost * Centre National de Génotypage, CEA - Institut de Génomique,
Evry, France Céline Besse & Jean-François Deleuze * Department of Pediatric Endocrinology, Bicêtre Hospital, Paris Sud University, Le Kremlin-Bicêtre, France Gianpaolo De Filippo &
Pierre Bougnères Authors * Delphine Fradin View author publications You can also search for this author inPubMed Google Scholar * Pierre-Yves Boëlle View author publications You can also
search for this author inPubMed Google Scholar * Marie-Pierre Belot View author publications You can also search for this author inPubMed Google Scholar * Fanny Lachaux View author
publications You can also search for this author inPubMed Google Scholar * Jorg Tost View author publications You can also search for this author inPubMed Google Scholar * Céline Besse View
author publications You can also search for this author inPubMed Google Scholar * Jean-François Deleuze View author publications You can also search for this author inPubMed Google Scholar *
Gianpaolo De Filippo View author publications You can also search for this author inPubMed Google Scholar * Pierre Bougnères View author publications You can also search for this author
inPubMed Google Scholar CONTRIBUTIONS D.F. performed sample preparations, carried out analyses and wrote the manuscript. P.Y.B. helped in the analysis. M.B.P. performed sample preparations.
F.L. and G.d.F. recruited cases and controls and provided blood samples. J.T. and C.B. ran D.N.A. methylation arrays. J.F.D., G.d.F. and P.B. provided intellectual input for the discussion
and critically revised the manuscript. D.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data
and the accuracy of the data analysis. All authors read and approved the final manuscript. CORRESPONDING AUTHOR Correspondence to Delphine Fradin. ETHICS DECLARATIONS COMPETING INTERESTS The
authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (PDF 52 KB) SUPPLEMENTARY DATASET (XLS 81 KB) RIGHTS AND PERMISSIONS This work is
licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license,
unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce
the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Fradin, D., Boëlle, PY., Belot,
MP. _et al._ Genome-Wide Methylation Analysis Identifies Specific Epigenetic Marks In Severely Obese Children. _Sci Rep_ 7, 46311 (2017). https://doi.org/10.1038/srep46311 Download citation
* Received: 23 November 2016 * Accepted: 14 March 2017 * Published: 07 April 2017 * DOI: https://doi.org/10.1038/srep46311 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative