Bacterial fermentation platform for producing artificial aromatic amines
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Aromatic amines containing an aminobenzene or an aniline moiety comprise versatile natural and artificial compounds including bioactive molecules and resources for advanced
materials. However, a bio-production platform has not been implemented. Here we constructed a bacterial platform for _para_-substituted aminobenzene relatives of aromatic amines _via_
enzymes in an alternate shikimate pathway predicted in a _Pseudomonad_ bacterium. Optimization of the metabolic pathway in _Escherichia coli_ cells converted biomass glucose to
4-aminophenylalanine with high efficiency (4.4 g L−1 in fed-batch cultivation). We designed and produced artificial pathways that mimicked the fungal Ehrlich pathway in _E. coli_ and
converted 4-aminophenylalanine into 4-aminophenylethanol and 4-aminophenylacetate at 90% molar yields. Combining these conversion systems or fungal phenylalanine decarboxylases, the
4-aminophenylalanine-producing platform fermented glucose to 4-aminophenylethanol, 4-aminophenylacetate and 4-phenylethylamine. This original bacterial platform for producing artificial
aromatic amines highlights their potential as heteroatoms containing bio-based materials that can replace those derived from petroleum. SIMILAR CONTENT BEING VIEWED BY OTHERS A BACTERIAL
PLATFORM FOR PRODUCING AROMATIC ESTERS FROM GLYCEROL Article 05 December 2024 A MICROBIAL PROCESS FOR THE PRODUCTION OF BENZYL ACETATE Article 23 February 2024 CELL-FREE PROTOTYPING ENABLES
IMPLEMENTATION OF OPTIMIZED REVERSE Β-OXIDATION PATHWAYS IN HETEROTROPHIC AND AUTOTROPHIC BACTERIA Article Open access 01 June 2022 INTRODUCTION The production of bulk materials and fuels
from renewable resources is prerequisite for constructing sustainable low-carbon societies that can overcome global environmental issues and limited fossil fuel resources. Aromatic amines
that are characterized by an amino-substituted benzene (aniline) moiety (referred to hereinafter as AA) serve as resources from which to develop dyes, rubbers, plastics and conductive
polymers1,2 and they are important in a broad range of industries. Most living organisms produce the AA, 4-aminobenzoic acid, as a biosynthetic precursor of folate, which is an essential
cofactor that is also a dietary supplement3. Some AA are intermediates of antibacterial chloramphenicol, pristinamycin4,5 and other drugs and developing a repertoire of AA is important from
a pharmaceutical standpoint. Due to such substantial demand, various commercial AA have been synthesized by petroleum chemistry, whereas none has been derived from biomass, which limits the
molecular design of practical bio-derived products based on AA. Aromatic amines are significant in the production of advanced polymer materials including functional and/or high-performance
plastics. The amine group and the aromatic moiety of AA induce nucleophilic reactivity and excellent thermomechanical performance, respectively6. Aromatic amines are polycondensed with
carbonyl compounds to generate aromatic polyamides, polyimides, polyazoles, polyurea and polyazomethines. When polycondensed with aromatic acids, AA generate super-engineering plastics with
extremely high thermomechanical properties7,8. These include poly(_p_-phenylene terephthalamide (KevlarTM) and poly(4,4′-oxydiphenylene pyromellitimide) (KaptonTM) that serve as thermostable
materials in fabric for body armor and other flame-retardant materials, fiber-reinforced plastics for electronic devices, vehicle bodies and anti-pressure cylinders7,8,9. The applications
of super-engineered plastics are diversifying and this is increasing the annual global production of AA-derived plastics to around 100,000 tons. Global production of aromatic polyamides
accounts for several hundreds of millions of US dollars, which indicates the size of the contribution of AA to both the economy and society. The production of aromatic polyamides and
polyimides requires aromatic carboxylic acids and/or AA as building blocks. Whereas aromatic carboxylic acids (such as terephthalic acid) have been derived from biomass10,11, AA have not and
this has precluded the development of fully bio-oriented aromatic polyamides and polyimides. Our recent microbial production of the non-natural AA, 4-aminocinnamic acid (4ACA), from
4-aminophenylalanine (4APhe), which is an intermediate of the chloramphenicol and pristinamycin biosynthesis pathway4,5, followed by synthesis of ultra-high-performance polyimide is the
exception12. Not only 4ACA, but also other AA derived from biomass would serve as innovative monomers for synthesizing bio-AA plastics and their environmental impact should be enormous,
considering that they would replace polyamides and polyimides derived in bulk from petroleum. Only a few natural pathways of AA biosynthesis have been documented. This causes considerable
difficulty when trying to custom-design pathways to synthesize various AA. The 4ACA production system ferments 4APhe _via_ antibiotic-synthesizing enzymes produced by recombinant
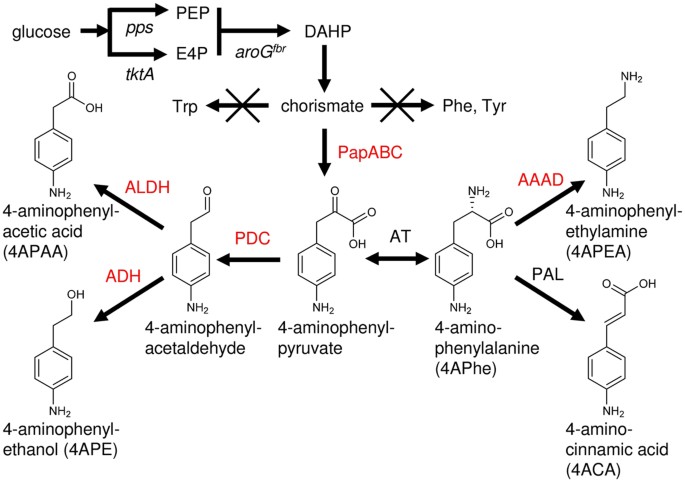
_Escherichia coli_13 and then a bacterium producing phenylalanine ammonia lyase converts 4APhe to 4ACA (Fig. 1)12. This study identified a novel 4APhe synthetic gene cluster from
_Pseudomonas fluorescens_, which in accordance with the optimization of host _E. coli_ metabolism enables 35-fold more 4APhe production than that generated in a previous study that used
antibiotic-synthesizing enzymes from _Streptomyces_ bacteria (0.13 g L−1)13. Recombinant _E. coli_ strains producing sets of phenylalanine-catabolic enzymes fermented glucose to a series of
artificial 4-amino-substituted AA including 4-aminophenylacetate (4APAA), 4-aminophenylacetaldehyde, 4-aminophenylethanol (4APE) and 4-aminophenylethylamine (4APEA) (Fig. 1). Bio-aromatic
polyamide and polyimides can be developed from all of these compounds. The platform for synthesizing bio-derived AA constructed in the present study increases the potential for producing a
range of bio-based AA materials for bio-polymers and other applications. RESULTS NOVEL _P. FLUORESCENS PAPABC_ GENES OPTIMIZE 4APHE PRODUCTION We and another group expressed the
_Streptomyces venezuelae_ and _S. pristinaespiralis_ genes encoding 4-amino-4-deoxychorismate synthase (PapA), 4-amino-4-deoxychorismate mutase (PapB) and 4-amino-4-deoxyprephenate
dehydrogenase (PapC) in recombinant _E. coli_ and synthesized 4APhe13. These enzymes convert cellular chorismate to 4-aminophenylpyruvate4,5, which endogenous aminotransferases in _E_.
_coli_ subsequently convert to 4APhe13. Homology searches identified similar amino acid sequences to _S. venezuelae_ PapA, PapB and PapC in predicted proteins of _P. fluorescens_ SBW25 (Fig.
2a) encoded by PFLU_1771 (_pfpapA_), PFLU_1772 (_pfpapB_) and PFLU_1770 (_pfpapC_) and their entire amino acid sequences have 44%, 28% and 34% similarity, respectively (Fig. 2a,
Supplementary Fig. 1). These genes are clustered in the _P. fluorescens_ SBW25 genome like their _Streptomyces_ counterparts4,5, where they might constitute an operon. We constructed an
artificial operon comprising the T7lac gene promoter followed by _pfpapB_, _pfpapA_ and _pfpapC_ in that order (plasmid 1 in Fig. 2a) and introduced it into _E. coli_ NST37(DE3)/Δ_pheLA_.
The strain lacks a _pheLA_ locus in the leader peptide for _pheA_ expression (_pheL_) and chorismate mutase (_pheA_) that competes with the substrate chorismate with PapA. The strain
harboring the operon generated 0.43 g L−1 4APhe in minimal medium with glucose whereas all strains expressing only _pfpapA_, _pfpapAB_ (Fig. 2b), _pfpapBC_, or the set of _pfpapA_ and
_pfpapC_ (Supplementary Fig. 2) did not. These results indicated that _pfpapABC_ participates in 4APhe synthesis. We analyzed the ability of a series of _pfpapABC_ expression plasmids to
produce 4APhe. The appropriate genes were introduced into pET-duet1, pRSF-duet1 and pCDF-duet1, which were maintained as ~40, 20~40 and >100 copies/cell, respectively14 and expressed
under the T7lac promoter (Fig. 2a). The strains harboring sets of pET-_pfpapA_ and pCDF-_pfpapB_C and pCDF-_pfpapA_ and pET-_pfpapBC_ (plasmids 3, 8 and 5, 6 in Fig. 2c, respectively)
produced the most 4APhe, up to 0.53 g L−1. We selected NST37(DE3)/Δ_pheLA_ harboring pET-_pfpapA_ and pCDF-_pfpapB_C (NDP strain), which produced 4APhe and optimized its 4APhe production.
METABOLIC ENGINEERING _E. COLI_ FOR EFFICIENT AA PRODUCTION We overexpressed the transketolase gene (_tktA_) under the control of the T7lac gene promoter in the NDP strain and found that it
produced 1.2-fold more 4APhe than the parent strain (Fig. 3a), which is consistent with reports that its overexpression increases the availability of erythrose 4-phosphate (E4P) for
3-deoxy-D-arabinoheptulosonate 7-phosphate (DAHP) synthase, the rate-limiting enzyme of chorismate production and hence cellular chorismate (Fig. 1)15,16 that is a substrate of the PapABC
pathway. The enzyme phosphoenolpyruvate (PEP) synthase is used to synthesize cellular PEP that is another substrate of shikimate pathway besides E4P. Overexpression of the PEP synthase gene
(_pps_) and both _tktA_ and _pps_ did not increase 4APhe production (Fig. 3a), indicating that _pps_ decreased 4APhe production under our culture conditions. The PEP-dependent carbohydrate:
phosphotransferase system (PTS) is a carbohydrate uptake process in _E. coli_ where it is the major consumer of intracellular PEP17. Aiming to eliminate PEP consumption by PTS during
cellular glucose uptake, we disrupted the _ptsHI-crr_ genes that encode the cytoplasmic components of the system. Introducing pET-_pfpapA_/pCDF-_pfpapB_C into the gene disruptant resulted in
lower 4APhe production compared with the corresponding non-disrupted strain (Supplementary Table 3), probably due to a decreased rate of glucose uptake. We overexpressed _aroG_ that encodes
DAHP synthase to provide more chorismate for 4APhe production (Fig. 1). We used the _aroG4__fbr_ gene that encodes a feedback inhibition-resistant (fbr) isozyme of DAHP synthase18. The NDPG
strain overexpressing _aroG4__fbr_ in the NDP strain produced 1.8-fold more 4APhe than the NDP strain in flask cultures (Fig. 3a) and this became more obvious when nitrogen sources in the
culture medium were increased (Supplementary Table 4). Therefore, the following tests included bacteria cultured under these conditions. Overexpressing _tktA_ and _pps_ did not increase
4APhe production by the NDPG strain (Supplementary Table 4), whereas it increased the production of phenylalanine by a strain sharing the same _aroG__fbr_ background15. These findings
indicate that the amount of DAHP synthase limits bacterial 4APhe production under these conditions. We fed-batch cultured NDPG in a jar fermenter and optimized the conditions for 4APhe
production. The glucose concentration decreased to <1 g L−1 after 16 h of batch culture and was maintained at this level thereafter to avoid repressing T7lac gene promoter activity19 and
the metabolic overflow of glucose to acetate that inhibits _E. coli_ fermentation20. Fed batch-cultured NDPG generated 4.4 g L−1 4APhe with a production yield of 17% (_vs._ glucose) (Fig.
3b), which represents 62% of the theoretical yield of phenylalanine by _E. coli_21. PRODUCTION OF 4APE AND 4APAA The fungal Ehrlich pathway deaminates phenylalanine to phenylpyruvate, which
phenylpyruvate decarboxylase (PDC) then converts to phenylacetaldehyde. Alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) reduce and oxidize phenylacetaldehyde to 2-phenylethanol
and to phenylacetic acid, respectively22. Artificial pathways mimicking the Ehrlich pathway were designed to produce 4APE and 4APAA (Fig. 1) _via_ pathway enzymes, although a
4-amino-substituted AA has never been assessed as an Ehrlich pathway intermediate. We constructed recombinant _E. coli_ BL21 Star (DE3) producing _ARO10_ encoding PDC from the fungus _S.
cerevisiae_22 and examined the bioconversion of 4APhe in resting cells. The reaction consumed 1.8 g L1 (10 mM) of 4APhe to generate 1.2 g L−1 of 4APE with a 90% molar yield (Table 1),
implying that PDC efficiently produced 4-aminophenylacetaldehyde, which endogenous _E. coli_ ADH reduced to 4APE. The resting _E. coli_ BL21 Star (DE3) cells producing both _ARO10_ and ALDH
encoding _ALD3_22 converted 1.8 g L−1 of 4APhe to 0.9 g L−1 of 4APAA with a 63% molar yield (Table 1). The strain produced 0.4 g L−1 4APE as a byproduct, indicating that the dehydrogenation
of 4-aminophenylacetaldehyde limits 4APAA production. Replacing _ALD3_ with _E. coli padA_23 and _S. cerevisiae ALD2_22 in the bacterium expressing _ARO10_ and _ALD3_ increased the yields of
4APAA to 85% and 90%, respectively (Table 1). We also combined PDC, _Pichia pastoris PpARO10_24 or _Aspergillus oryzae ppdA_25 with _ALD3_, but they produced less 4APAA than those
expressing _ARO10_ and _ALD3_ (Table 1). These results indicated that the heterologous expression of _ARO10_ alone and of the _ARO10_ and _ALD2_ set converted 4APhe to 4APE and APAA the most
efficiently. Figure 4a,b shows the time-dependent bioconversion of 4APhe (3.6 g L−1) in the optimized systems that produced 2.8 g L−1 4APE and 2.7 g L−1 4APAA. We expressed _ARO10_ or both
_ARO10_ and _ALD2_ in the NDPG strain expressing _pfpapABC_. The resulting NDPGA and NDPGAA strains cultured in fermentation medium produced 4APE and both 4APAA and 4APE, respectively.
Production increased linearly until reaching a maximum of 0.24 g L−1 4APE (Fig. 4c), 0.12 g L−1 4APAA and 0.19 g L−1 4APE (Fig. 4d) at 36 h. These results showed that the cultures fermented
glucose to 4APE and 4APAA. Both strains accumulated less fermentation products than the intermediate, 4APhe (0.4–1.2 g L−1) (Fig. 4c,d). Further optimization of growth parameters or genetic
modification could hasten conversion of the intermediate and produce more 4APE and 4APAA. Fermentation by the NDPGAA generated 4APE as a byproduct (Fig. 4d), while bioconversion by _E. coli_
BL21 Star (DE3) expressing _ARO10_ and _ALD3_ resulted in little 4APE production (Fig. 4b), indicating metabolic differences between the strains. These results demonstrated that the new
bacterial platform can efficiently produce biomass-derived AA. PRODUCTION OF 4APEA We examined the reaction of aromatic L-amino acid decarboxylase, which decarboxylates phenylalanine to
phenylethylamine and carbon dioxide, against 4APhe (Fig. 1). We constructed recombinant _E. coli_ BL21 Star (DE3) overexpressing _A. oryzae aadA_25 and either of the _Solanum lycopersicum
LeAADC1A_ and _LeAADC1B_26 genes, both of which encode aromatic L-amino acid decarboxylases. Incubating these cells with 1.8 g L−1 (10 mM) 4APhe efficiently converted 4APhe to 4APEA with 65%
to 75% molar yield (_vs._ 4APhe) (Fig. 5a). The strain expressing either _LeAADC1A_ or _LeAADC1B_ produced 4APEA within 4 h. We introduced these genes into the NDPG strain and cultured it
in fermentation medium containing 1% glucose. After 42 h, 1.3-fold more 4APEA was accumulated by the strain expressing _LeAADC1A_ than _LeAADC1B_ (Fig. 5b), indicating that both strains
harboring _LeAADC1A_ converted glucose biomass to 4APEA more efficiently. Fed-batch cultures of the strain expressing _LeAADC1A_ in a jar fermenter generated 1.8 g L−1 4APEA after 44 h (Fig.
5c). The production yield (_vs._ glucose) of 4APEA was 5.6%, which was below its theoretical yield (23%) calculated from that for phenylalanine (27.5%)21. DISCUSSION The aim of constructing
microbial platforms to produce various groups of chemicals is to develop specific pharmaceuticals and biomass-derived materials that can substitute for petroleum chemicals. Most current
targets for petroleum-based chemicals comprise aliphatic molecules, whereas only aromatic amino acids are fermented using glucose biomass as a carbon donor. Glucose is bio-converted into
shikimate pathway intermediates27 and deoxy-_scyllo_-inosose28, both of which are chemically derivatized to aromatic compounds such as catechol and its related compounds. Bio-derived
terephthalic acid has recently been synthesized using biomass-derived isobutanol or 5-hydroxymethylfurfral11. These aromatic compounds were generated by combining microbial, with chemical
reactions. However, we explored biological production systems that do not require chemical processes to produce artificial aromatic chemicals, especially those with a substituted nitrogen
atom. This is the first bacterial platform for the direct fermentative production of a series of AA and represents a promising environmentally-friendly alternative for synthesizing AA
building blocks that are applicable to numerous technologies. This study integrated metabolic engineering strategies to produce AA. Firstly, established strategies for high-level
fermentation of phenylalanine as well as perturbed metabolic flow of chorismate to phenylalanine improved chorismate synthesis by _E. coli._ Secondly, introducing exogenous _papABC_ genes
integrated the intrinsic _E. coli_ pathway for chorismate synthesis with the pathway generating aminophenylpyruvate from chorismate and fermented glucose to 4APhe (Fig. 1). We found that
_papABC_ is indispensable for AA production and that the _P. fluorescens papABC_ identified herein and which originated from a proteobacterium similar to _E. coli_ optimized production
efficiency. Thirdly, we searched bacteria, fungi and plants for enzymes that produce shikimate pathway derivatives and used them to produce 4APEA, 4APE and 4APAA from a sugar biomass. The
production rates are sufficient to generate industrial quantities of pharmaceutical intermediates. Fed-batch fermentation produced optimal amounts of 4APEA (1.8 g L−1), which will be
improved by applying traditional strategies such as continuous culture and genetically engineering host _E. coli_ metabolism and 4APEA generated in this manner will have potential for
bio-material production after a purification process is established. The platform produced biomass-derived aromatics containing an aniline structure with a substitution at the
_para_-position _via_ a C2 chain linked to a functional (amine or carbonyl) group. Such structures have unique molecular properties; 4APEA is a diamine connected to rigid aromatic (C6)
moiety and flexible C2 chain. This and structurally similar bipartite diamines are polymerized with both aliphatic and aromatic dicarboxylic acids to generate thermostable polyamides with a
considerably high glass-transition temperature29. Polyamide generated from 4APEA and isophthalic acid has high thermal stability (_T_d10 = 360 °C)30. Polycondensation with long flexible
aliphatic acids generates 4APEA with liquid crystallinity. Furthermore, 4APAA is a C6:C2 compound and an aromatic amino acid. The related 4-aminobenzoic acid (C6:C1) is homopolymerized to
form poly(_p_-benzamide) with high thermomechanical properties but its poor solubility and the absence of a melting temperature causes difficulties with processing them into plastics.
Copolymerizing 4-aminobenzoic acid with 4APAA increases flexibility through the addition of alkyl chains connected to the aromatic rings of poly(_p_-benzamide)31. This molecular design
improves processability into heat-resistant filaments, fibers and films31. Thus, the present findings should impact the development of novel bio-based aromatic materials. We developed an AA
production platform based on a shikimate pathway variant for synthesizing 4APhe (Fig. 1). Although only a small variety of compounds are derived from this pathway in nature, the platform
produced the artificial 4-amino-substituted compounds 4APAA, 4APE and 4APAE in combination with the enzymes for synthesizing their phenylalanine derivatives. Natural derivatives of
phenylalanine and its synthetic intermediates are more diverse than 4APhe-related compounds and include cinnamic acid, cinnamyl alcohol, cinnamaldehyde, homogentisic acid, phenyllactic acid,
mandelic acid, styrene, benzoic acid and other aromatic compounds32. We use phenylalanine-deaminating phenylalanine ammonia lyase (Fig. 1) in the bioconversion of 4APhe to 4ACA11. Other
enzymes that biosynthesize phenylalanine derivatives are potential catalysts in the generation of artificial AA, but their synthesis awaits investigation. The constructed platform for AA
synthesis has the potential to produce bio-derived non-natural AA. METHODS BACTERIAL STRAINS AND REAGENTS Supplementary Table 1 lists the strains used in this study. _Escherichia coli_ NST37
(ATCC31882) was lysogenized using λDE3 Lysogenization kits (Novagen, Madison, WI, USA) to generate NST37 (DE3). We obtained 4APhe from Sigma Aldrich (St. Louis, MO, USA) and 4ACA, 4APE and
4APEA from Tokyo Chemical Industry (Tokyo, Japan). Plasmids were constructed using PrimeSTAR HS DNA polymerase and restriction enzymes (Takara Bio Inc., Shiga, Japan). CONSTRUCTING PLASMIDS
FOR 4APHE PRODUCTION Supplementary Table 2 lists the primers used in this study. Artificially synthesized, codon-optimized _pfpapA_, _pfpapB_ and _pfpapC_ (GenScript, NJ, USA) (accession
numbers; KU199222, KU199223 and KU199224) were cloned into pUC57 to generate pUC-pfpapA, pUC-pfpapB and pUC-pfpapC, respectively. DNA fragments for _pfpapA_, _pfpapB_ and _pfpapC_ were
amplified by PCR using these plasmids and the respective primers sets, PFAF/PFAR, PFBF/PFBR and PFCF/PFCR and then fused by PCR using the primers PFBF and PFCR to generate _pfpapBAC_
fragments. These fragments were digested with _Nde_I and _Xho_I and cloned into pET22b to generate pET-pfpapBAC. Fragments of pfpapBA were amplified by PCR using PFBF and PFAR2 primers and
pET-pfpapBAC, digested with _Nde_I and _Xho_I and cloned into pET22b to generate pET-pfpapBA. We digested pUC-pfpapA with _Nde_I and _Xho_I and then purified _pfpapA_ fragments were cloned
into pET-duet1, pCDF-duet1 and pRSF-duet-1 (Novagen) that were digested with the same enzymes to generate pET-pfpapA, pCDF-pfpapA and pRSF-pfpapA. We digested pUC-pfpapB with _Nco_I and
_Not_I and inserted them into the same vectors to generate pET-pfpapB, pCDF-pfpapB and pRSF-papB. We digested pUC-pfpapC with _Nde_I and _Xho_I and cloned _pfpapC_ fragments into these
plasmids to generate pET-pfpapBC, pCDF-pfpapBC and pRSF-papBC. GENETIC ENGINEERING OF INTRINSIC _E. COLI_ METABOLIC PATHWAYS The synthesized DNA fragment _aroG4__fbr_ 18 was cloned into
pACYC184 that was digested with _Eco_RV and _Hi_ndIII and treated with the Klenow fragment to create pACYC-aroG4. The _tktA_ and _pps_ genes were amplified by PCR using _E. coli_ MG1655
total DNA and primers, digested with _Nco_I and _Bam_HI and _Nde_I and _Xho_I and cloned into pRSFduet-1 to generate pRSF-tktA and pRSF-pps, respectively. The _pps_ gene was digested with
_Nde_I and _Xho_I and also cloned into pRSF-tktA to generate pRSF-tktApps. Knock-out cassettes generated by PCR using the primers DpheLAF and DpheLAR and the Red/ET recombination system
(Gene Bridges, Heidelberg, Germany) replaced the genomic _pheLA_ locus of _E. coli_ NST37(DE3) with the kanamycin resistance gene (Kmr). The FLP/FRT recombination technique (Gene Bridges)
deleted Kmr from the strain and generated _E. coli_ NST37(DE3)/Δ_pheLA_. Diagnostic PCR proceeded using the primers C1 and C2. Likewise, the _ptsHI_-_crr_ genes of NST37(DE3)/Δ_pheLA_ were
knocked-out using the cassette-generated primer pairs DptsHI-crrF and DptsHI-crrR to generate NST37(DE3)/Δ_pheLA_/Δ_ptsHI-crr_. Primer pairs C3 and C4 was used for diagnostic PCR.
CONSTRUCTING PLASMIDS FOR PRODUCTION OF OTHER AA The _ARO10_ and _ALD3_ genes were amplified by PCR using total DNA from _S. cerevisiae_ and appropriate primers. The _ARO10_ fragment was
digested with _Nco_I and _Bam_HI and cloned into pRSFduet-1 to generate pRSF-aro10. The ALD3 fragment was digested with _Nde_I and _Xho_I and cloned into pRSFduet-1 and pRSF-aro10 to
generate pRSF-ald3 and pRSF-aro10ald3, respectively. The _S. cerevisiae ALD2_ and the _E. coli padA_ genes were amplified using the appropriate primers, digested with _Nde_I and _Xho_I and
cloned into pRSF-aro10 to generate pRSF-aro10ald2 and pRSF-aro10padA. The PpARO10 (GenBank accession number, CCA40086.1) gene was amplified by PCR using _Pichia pastoris_ total DNA and
primers, digested with _Sac_I and _Not_I and then cloned into pRSF-ald3 to generate pRSF-pparo10ald3. The _A. oryzae ppdA_ was amplified using primers, digested with _Nco_I and _Hin_dIII and
cloned into pRSFduet-1 to generate pRSF-ppdA. The _Nde_I and _Xho_I fragment of _ALD3_ was cloned into pRSF-ppdA to generate pRSF-ppdAald3. _Solanum lycopersicum_ LeAADC1A and LeAADC1B were
amplified using its cDNA and the appropriate primers. The _A. oryzae aadA_ gene was amplified using pET-aadA and the appropriate primers. These DNA fragments were digested with _Bam_HI and
_Hin_dIII and cloned into pRSFduet-1 to generate pRSF-leaadc1a, pRSF-leaadc1b and pRSF-aadA. FERMENTATION OF 4APHE, 4APAA, 4APE AND 4APEA Cells were grown in the fermentation medium
comprising 10 g glucose, 2 g tryptone, 1 g yeast extract, 6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 2 g NH4Cl, 0.5 g MgSO4 7H2O, 15 mg CaCl2, 20 mg tyrosine, 20 mg tryptophan, 20 mg
phenylalanine, 50 mg thiamine-HCl and 1 mL of trace element solution/L33. The plasmids were maintained by adding 100 mg L−1 sodium ampicillin, 40 mg L−1 kanamycin sulfate and 35 mg L−1
chloramphenicol. Recombinant _E. coli_ generated from NST37(DE3)/Δ_pheLA_ was pre-cultured by rotary shaking at 300 rpm overnight in test tubes containing 5 mL of LB medium at 28 °C and
inoculated into 100 mL of fresh M9 medium in 500-mL conical flasks at 1:100 dilution. When the cells reached an optical density (OD) of 0.6 at 600 nm and 30 °C, 0.1 mM
isopropyl-β-d-thiogalactoside (IPTG) was added. After 20–24 h of cultivation the glucose concentration reached <2 g L−1 and then 2 mL of 500 g L−1 glucose was added and the cells were
further cultured in the flasks for 48 h. Fed-batch cultures were agitated at 30 °C and 550 rpm in a 1.0-L BMJ-01 fermenter (Biott, Tokyo, Japan) containing 0.5 L of fermentation medium
supplemented with 5 g L−1 tryptone, 2.5 g L−1 yeast extract, 10 g L−1 (NH4)2SO4 and 10 g L−1 glucose. The culture was aerated at 0.6 L min−1. When the OD reached 0.6 at 600 nm, 0.1 mM IPTG
was added. Peristaltic pumps fed the cultures with 500 g L−1 of glucose and 0.1 mM IPTG when the glucose concentration dipped below 1.5 g L−1. The pH was monitored using an electrode and
maintained between 7.0 and 7.1 by adding 10% NH4OH. BIOCONVERSION BY RESTING CELLS _Escherichia coli_ BL21 Star (DE3) (Invitrogen, Carlsbad, CA, USA) harboring expression plasmids was rotary
shaken at 300 rpm and 30 °C overnight in test tubes containing 3 mL of LB medium and inoculated into 500-mL conical flasks containing 100 mL LB medium at a 1:100 dilution. When the cells
reached an OD of 0.6 at 600 nm and 30 °C, 0.1 mM IPTG was added. Cells harboring pRSF-aro10, pRSF-aro10ald3, pRSF-aro10ald2 or pRSF-aro10padA were further incubated at 30 °C for 6 h. The
cells were collected by centrifugation at 5,000 × _g_ for 10 min and washed with DCD buffer (100 mM KH2PO4 (pH 7.5), 1 mM MgSO4, 0.5 mM thiamine chloride). Cells harboring pRSF-leaadc1a,
pRSF-leaadc1b or pRSF-aadA were further incubated at 16 °C for 12 h with 0.5 mM pyridoxal 5-phosphate, washed with AAD buffer (100 mM potassium phosphate, pH 7.5 and 0.5 mM pyridoxal
5-phosphate). The washed cells were incubated in 10 mL of DCD or AAD buffer containing the indicated amounts of 4APhe at 28 °C and shaken at 300 rpm for the indicated periods. DETERMINATION
OF METABOLITES We determined yields of 4APhe, 4APA, 4APE and 4APEA by high-performance liquid chromatography (HPLC) using a 1200 infinity series (Agilent Technologies, Palo Alto, CA, USA)
equipped with a 250 × 4.6-mm Purospher Star RP-18 end-capped column with a particle size of 5 μm (Millipore-Merck, Billerica, MA, USA). The initial mobile phase was solvent A:solvent B =
98:2 (solvent A, 20 mM potassium phosphate (pH 7.0); solvent B, methanol) and maintained for 7 min. The concentration of solvent B was increased to 50% for 5 min and then maintained at that
ratio for another 5 min. The flow rate was 0.8 mL min−1 and absorption at 210 nm was monitored. Glucose concentrations were determined using glucose-CII test kit (Wako, Tokyo, Japan).
ADDITIONAL INFORMATION HOW TO CITE THIS ARTICLE: Masuo, S. _et al._ Bacterial fermentation platform for producing artificial aromatic amines. _Sci. Rep._ 6, 25764; doi: 10.1038/srep25764
(2016). REFERENCES * Lawrence, A. S. Amines: synthesis, properties and applications. Cambridge University Press (2006). * Sousa, A. C., Martins, L. O. & Robalo, M. P. Laccase-catalysed
homocoupling of primary aromatic amines towards the biosynthesis of dyes. Adv. Synth. Catal. 355, 2908–2917 (2013). Article CAS Google Scholar * Krishnaswamy, K. & Madhavan Nair, K.
Importance of folate in human nutrition. Br. J. Nutr. 85, S115–124 (2001). Article CAS Google Scholar * He, J., Magarvey, N., Piraee, M. & Vining, L. C. The gene cluster for
chloramphenicol biosynthesis in _Streptomyces venezuelae_ ISP5230 includes novel shikimate pathway homologues and a monomodular non-ribosomal peptide synthetase gene. Microbiol. 147,
2817–2829 (2001). Article CAS Google Scholar * Blanc, V. et al. Identification and analysis of genes from _Streptomyces pristinaespiralis_ encoding enzymes involved in the biosynthesis of
the 4-dimethylamino-L-phenylalanine precursor of pristinamycin I. Mol. Microbiol. 23, 191–202 (1997). Article CAS ADS Google Scholar * Fink, J. K. High Performance Polymers. William
Andrew Inc. (2008). * Liaw, D. J. et al. Advanced polyimide materials: Syntheses, physical properties and applications. Prog. Polym. Sci. 37, 907–974 (2012). Article CAS Google Scholar *
García, J. M., García, F. C., Serna, F. & de la Peña, J. L. High-performance aromatic polyamides. Prog. Polym. Sci. 35, 623–686 (2010). Article Google Scholar * Sapurina, I. The
mechanism of the oxidative polymerization of aniline and the formation of supramolecular polyaniline structures. Polym. Int. 57, 1295–1325 (2008). Article CAS Google Scholar * Pacheco, J.
J. & Davis, M. E. Synthesis of terephthalic acid via Diels-Alder reactions with ethylene and oxidized variants of 5-hydroxymethylfurfural. Proc. Natl Acad. Sci. USA 111, 8363–8367
(2014). Article CAS ADS Google Scholar * Lin, Z., Nikolakis, V. & Ierapetritou, M. Alternative approaches for _p_-xylene production from starch: techno-economic analysis. Ind. Eng.
Chem. Res. 53, 10688–10699 (2014). Article CAS Google Scholar * Suvannasara, P. et al. Biobased polyimides from 4-aminocinnamic acid photodimer. Macromolecules 47, 1586–1593 (2014).
Article CAS ADS Google Scholar * Mehl, R. A. et al. Generation of a bacterium with a 21 amino acid genetic code. J. Am. Chem. Soc. 125, 935–939 (2003). Article CAS Google Scholar *
Tolia, N. H. & Joshua-Tor, L. Strategies for protein coexpression in _Escherichia coli_. Nat. Methods 3, 55–64 (2006). Article CAS Google Scholar * Patnaik, R. & Liao, J. C.
Engineering of _Escherichia coli_ central metabolism for aromatic metabolite production with near theoretical yield. Appl. Environ. Microbiol. 60, 3903–3908 (1994). CAS PubMed PubMed
Central Google Scholar * Sprenger, G. A. From scratch to value: engineering _Escherichia coli_ wild type cells to the production of L-phenylalanine and other fine chemicals derived from
chorismate. Appl. Microbiol. Biotechnol. 75, 739–749 (2007). Article CAS Google Scholar * Rodriguez, A. et al. Engineering _Escherichia coli_ to overproduce aromatic amino acids and
derived compounds. Microb. Cell Fact. 13, 126 (2014). PubMed PubMed Central Google Scholar * Kikuchi, Y., Tsujimoto, K. & Kurahashi, O. Mutational analysis of the feedback sites of
phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase of _Escherichia coli_. Appl. Environ. Microbiol. 63, 761–762 (1997). CAS PubMed PubMed Central Google Scholar
* Grossman, T. H., Kawasaki, E. S., Punreddy, S. R. & Osburne, M. S. Spontaneous cAMP-dependent derepression of gene expression in stationary phase plays a role in recombinant
expression instability. Gene 209, 95–103 (1998). Article CAS Google Scholar * Gerigk, M. et al. Process control for enhanced L-phenylalanine production using different recombinant
_Escherichia coli_ strains. Biotechnol. Bioeng. 80, 746–754 (2002). Article CAS Google Scholar * Förberg, C., Eliaeson, T. & Häggström, L. Correlation of theoretical and experimental
yields of phenylalanine from non-growing cells of a rec _Escherichia coli_ strain. J. Biotechnol. 7, 319–331 (1988). Article Google Scholar * Kim, B., Cho, B. R. & Hahn, J. S.
Metabolic engineering of _Saccharomyces cerevisiae_ for the production of 2-phenylethanol via Ehrlich pathway. Biotechnol. Bioeng. 111, 115–124 (2014). Article CAS Google Scholar *
Ferrandez, A., Prieto, M. A., Garcia, J. L. & Diaz, E. Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from _Escherichia coli_. FEBS Lett. 406, 23–27 (1997).
Article CAS Google Scholar * Küberl, A. et al. High-quality genome sequence of _Pichia pastoris_ CBS7435. J. Biotechnol. 154, 312–320 (2011). Article Google Scholar * Masuo, S., Osada,
L., Zhou, S., Fujita, T. & Takaya, N. _Aspergillus oryzae_ pathways that convert phenylalanine into the flavor volatile 2-phenylethanol. Fungal Genet. Biol. 77, 22–30 (2015). Article
CAS Google Scholar * Tieman, D. et al. Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl Acad.
Sci. USA 103, 8287–8292 (2006). Article CAS ADS Google Scholar * Li, W., Xie, D. & Frost, J. W. Benzene-free synthesis of catechol: interfacing microbial and chemical catalysis. J.
Am. Chem. Soc. 127, 2874–2882 (2005). Article CAS Google Scholar * Kogure, T., Wakisaka, N., Takaku, H. & Takagi, M. Efficient production of 2-deoxy-scyllo-inosose from d-glucose by
metabolically engineered recombinant _Escherichia coli_. J. Biotechnol. 129, 502–509 (2007). Article CAS Google Scholar * Lee, L. T. C. High-transition-temperature polyamides based on
2(2-aminophenyl)1,1-dimethylethylamine. J. Polym. Sci. 16, 2025–2038 (1978). CAS Google Scholar * Ueda, M., Morishima, M. & Kakuta, M. Synthesis of sequential polyamide by direct
polycondensation II. Polym. J. 23, 1511–1517 (1991). Article CAS Google Scholar * Wang, H. H. & Lin, M. F. Modification of nylon-6 with wholly rigid poly(m-phenylene isophthalamide).
J. Appl. Polym. Sci. 43, 259–269 (1991). Article CAS Google Scholar * Thompson, B., Machas, M. & Nielsen, D. R. Creating pathways towards aromatic building blocks and fine chemicals.
Curr. Opin. Biotechnol. 36, 1–7 (2015). Article CAS Google Scholar * Hutner, S. H. Anaerobic and aerobic growth of purple bacteria (athiorhodaceae) in chemically defined Media. Microbiol.
4, 286–293 (1950). CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank Norma Foster for critical reading of the manuscript. We thank Drs. Ariizumi, T. and Ezura, H.
(University of Tsukuba) for providing _S. lycopersicum_ cDNA. This work was supported by Advanced Low Carbon Technology Research and Development Program (5100270) and CREST from the Japan
Science and Technology Agency. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Faculty of Life and Environmental Sciences, University of Tsukuba, Tsukuba, 305-8572, Ibaraki, Japan Shunsuke
Masuo, Shengmin Zhou & Naoki Takaya * School of Materials Science, Japan Advanced Institute of Science and Technology, 1-1 Asahidai, Nomi, 923-1292, Ishikawa, Japan Tatsuo Kaneko Authors
* Shunsuke Masuo View author publications You can also search for this author inPubMed Google Scholar * Shengmin Zhou View author publications You can also search for this author inPubMed
Google Scholar * Tatsuo Kaneko View author publications You can also search for this author inPubMed Google Scholar * Naoki Takaya View author publications You can also search for this
author inPubMed Google Scholar CONTRIBUTIONS S.M. and N.T. designed research and analyzed data. S.M., T.K. and N.T. wrote the paper. S.M. and S.Z. performed the experiments. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY INFORMATION RIGHTS AND PERMISSIONS This work is
licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license,
unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce
the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Masuo, S., Zhou, S., Kaneko, T.
_et al._ Bacterial fermentation platform for producing artificial aromatic amines. _Sci Rep_ 6, 25764 (2016). https://doi.org/10.1038/srep25764 Download citation * Received: 22 December 2015
* Accepted: 21 April 2016 * Published: 11 May 2016 * DOI: https://doi.org/10.1038/srep25764 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content:
Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative