Patient-derived tumor organoids for personalized medicine in a patient with rare hepatocellular carcinoma with neuroendocrine differentiation: a case report
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT BACKGROUND Hepatocellular carcinoma with neuroendocrine differentiation (HCC-NED) is a very rare subtype of primary liver cancer. Treatment allocation in these patients therefore
remains a challenge. METHODS We report the case of a 74-year-old man with a HCC-NED. The tumor was surgically removed in curative intent. Histopathological work-up revealed poorly
differentiated hepatocellular carcinoma (Edmondson-Steiner grade IV) with diffuse expression of neuroendocrine markers synaptophysin and chromogranin. Three months after resection,
multifocal recurrence of the HCC-NED was observed. In the meantime, tumor organoids have been generated from the resected HCC-NED and extensively characterized. Sensitivity to a number of
drugs approved for the treatment of HCC or neuroendocrine carcinomas was tested in vitro. RESULTS Based on the results of the in vitro drug screening, etoposide and carboplatin are used as
first line palliative combination treatment. With genomic analysis revealing a _NTRK1_-mutation of unknown significance (kinase domain) and tumor organoids found to be sensitive to
entrectinib, a pan-TRK inhibitor, the patient was treated with entrectinib as second line therapy. After only two weeks, treatment is discontinued due to deterioration of the patient’s
general condition. CONCLUSION The rapid establishment of patient-derived tumor organoids allows in vitro drug testing and thereby personalized treatment choices, however clinical translation
remains a challenge. To the best of our knowledge, this report provides a first proof-of-principle for using organoids for personalized medicine in this rare subtype of primary liver
cancer. PLAIN LANGUAGE SUMMARY Tumors that simultaneously display features of liver, nerve and hormone-producing cells are very rare. In such cases, the most appropriate treatment choice is
not well defined. Here, we describe the generation of three-dimensional miniature tumors, called organoids, from the patient’s tumor tissue, that can be grown and studied in a culture dish.
These organoids closely mimic the patient’s tumor and allowed us to test different drugs to identify the most effective therapy for informed treatment choice. What we describe in this study
is an emerging approach for a practice known as personalized medicine, that aims to provide a more tailored treatment to patients. In summary, we demonstrate that this approach can be useful
in a rare cancer type and that it holds significant potential to guide treatment decision in other patients with aggressive cancers. SIMILAR CONTENT BEING VIEWED BY OTHERS DRAMATIC, DURABLE
RESPONSE TO THERAPY IN G_BRCA2_-MUTATED PANCREAS NEUROENDOCRINE CARCINOMA: OPPORTUNITY AND CHALLENGE Article Open access 22 April 2023 REPID AS A POTENTIAL BIOMARKER AND THERAPEUTIC TARGET
FOR LUNG NEUROENDOCRINE TUMOR Article Open access 11 November 2024 MOLECULAR PATHOGENESIS AND SYSTEMIC THERAPIES FOR HEPATOCELLULAR CARCINOMA Article 28 April 2022 INTRODUCTION Primary liver
carcinomas with concurrent hepatocellular and neuroendocrine tumor components in the same liver lesion are very rare1. They consist of two morphologically distinct cell populations that
express hepatocellular or neuroendocrine markers and are classified as Hepatocellular Carcinoma-Neuroendocrine Carcinoma (HCC-NEC)2 or liver mixed neuroendocrine non-neuroendocrine neoplasms
(MiNEN)3. The published case reports describe an aggressive tumor phenotype and poor overall prognosis3,4. Even rarer are HCCs with neuroendocrine differentiation (HCC-NED)5. HCC-NEDs are
comprised of morphologically uniform cells that stain positively for both hepatocellular and neuroendocrine markers. Patients are usually treated by means of surgical resection,
transarterial chemoembolization or systemic (chemo)therapy for liver cancer or neuroendocrine malignancies. Because HCC with neuroendocrine differentiation is a very rare tumor entity,
therapy in these patients remains ill-defined4,6,7. Here, we report a case history of a 74-year-old man with HCC-NED. We provide a comprehensive histopathological characterization and a
genomic analysis of this rare tumor. Furthermore, we describe the generation of tumor organoids that retain the key characteristics of the originating tumor. The organoids were used in drug
screens to identify the most promising treatment options. METHODS PATIENT INFORMATION AND BIOLOGICAL MATERIAL Human biopsy and resection tissue was collected from patients undergoing
diagnostic liver biopsy or liver surgery at the University Hospital of Basel. Written informed consent was obtained from all patients. The study was approved by the local ethics committee
(protocol numbers EKNZ 2014-099 as well as BASEC 2019-02118). For the HCC-NED patient described in this study, written informed consent to publish the case details was obtained from the
family. LIVER CANCER ORGANOID CULTURE Tumor organoid lines were generated from liver biopsy or resection tissue according to published protocols8,9. Briefly, tumor tissues were dissociated
to small-cell clusters and seeded in domes of basement membrane extract type 2 (BME2, R&Dsystems, Cat. No. 3533-005-02). Polymerized BME2 domes were overlaid with expansion medium (EM):
advanced DMEM/F-12 (Gibco, Cat. No. 12634010) supplemented with 1× B-27 (Gibco, Cat. No. 17504001), 1× N-2 (Gibco, Cat. No. 17502001), 10 mM Nicotinamide (Sigma, Cat. No. N0636), 1.25 mM
N-Acetyl-L-cysteine (Sigma, Cat. No. A9165), 10 nM [Leu15]-Gastrin (Sigma, Cat. No. G9145), 10 μM Forskolin (Tocris, Cat. No. 1099), 5 μM A83-01 (Tocris, Cat. No. 2939), 50 ng/ml EGF
(Peprotech, Cat. No. AF-100-15), 100 ng/ml FGF10 (Peprotech, Cat. No. 100-26), 25 ng/ml HGF (Peprotech, Cat. No. 100-39), 10% RSpo1-conditioned medium (v/v, homemade). Cultures were kept at
37 °C in a humidified 5% CO2 incubator. Organoids were passaged weekly at 1:4–1:6 split ratios using 0.25% Trypsin-EDTA (Gibco, Cat. No. 25200056). Frozen stocks were prepared at regular
intervals. All organoid cultures were regularly tested for Mycoplasma contamination using the MycoAlert™ Mycoplasma detection kit (Lonza, Cat. No. LT07-118). HISTOLOGY AND
IMMUNOHISTOCHEMISTRY Tumor and liver tissues were fixed in 4% phosphate-buffered formalin and embedded in paraffin using standard procedures. Tumor organoids were released from BME2 by
incubation in Dispase II (Sigma-Aldrich, Cat. No. D4693). Organoids were fixed in 4% phosphate-buffered formalin in PBS for 30 min at room temperature following encapsulation in HistoGel
(Thermo Fisher Scientific, Cat. No. HG-4000-012) and subsequent dehydration and paraffin embedding. Histopathological evaluation was assessed by three board-certified pathologists (MSM, JV
and LMT). Tumors were classified based on architecture and cytological features, and graded according to the Edmondson grading system10,11. The following primary antibodies were used for
automated diagnostic immunohistochemical staining on a Benchmark XT device (Ventana Medical Systems) at the Institute of Pathology of the University of Basel: AFP (Ventana, Ref-Nr.
760-2603), ARG1 (Ventana, Ref-Nr. 760-4801), CD10 (Ventana, Ref-Nr. 790-4506), CD56 (Ventana, Ref-Nr. 790-4465), CHGA (Ventana, Ref-Nr. 760-2519), GPC3 (Ventana, Ref-Nr. 790-4564), HLA-ABC
(Abcam, Cat. No. ab70328), Hep Par-1 (Ventana, Ref-Nr. 760-4350), KRT19 (Ventana, Ref-Nr. 760-4281), Ki-67 (Dako, Cat. No. IR626), Pan-TRK (Abcam, Cat. No. ab181560), PD-L1 (Ventana, Ref-Nr.
740-4907), SYP (Ventana, Ref-Nr. 790-4407), and SSTR2 (Abcam, Cat. No. ab134152). XENOGRAFT MOUSE MODEL Experiments involving animals were performed in strict accordance with Swiss law and
were previously approved by the Animal Care Committee of the Canton Basel-Stadt, Switzerland. Tumor organoids, corresponding to 2 × 106 cells, were released from BME2, resuspended in 100 μl
50:50 (v/v) BME2:expansion medium, and injected subcutaneously into the flank of one male NSG (Non-obese diabetic, Severe combined immunodeficiency, Gamma) mouse (The Jackson Laboratory) at
8 weeks of age. The mouse was housed in an individually ventilated cage (Tecniplast Green Line) at 22 °C, 55% humidity and a light cycle of 12:12 h. Tumor growth was assessed weekly by
caliper measurement. The tumor was harvested when it reached 1000 mm3 in size, fixed in 4% phosphate-buffered formalin and processed for paraffin embedding and immunohistochemistry as
described above. DRUG SCREENINGS All compounds were dissolved in DMSO at 10 mM (except for cisplatin and carboplatin) and aliquots were stored at −20 °C, 4 °C or room temperature according
to the manufacturer’s recommendations. Sorafenib tosylate, lenvatinib mesylate, cabozantinib mesylate, regorafenib, octreotide acetate, lanreotide acetate, etoposide, sunitinib malate,
everolimus, entrectinib, larotrectinib: all from Selleckchem; pasireotide ditrifuloroacetate (MedChem Express); cisplatin (Sandoz); carboplatin (Labatec). For drug screenings, organoids were
dissociated with 0.25% Trypsin-EDTA (Gibco) and seeded at 1000 cells/well in 384-well plates in organoid expansion medium supplemented with 10% BME2. Two days later, compounds were added in
a 2-fold dilution series ranging from 0.02 nM to 10 μM. After 6 days of treatment, cell viability was measured using CellTiter-Glo 3D (Promega). Luminescence was measured on a Synergy H1
Multi-Mode Reader (BioTek Instruments). Results were normalized to vehicle control (100% DMSO or 100% water). All experiments were performed twice. Dose-response curves were calculated using
Prism 9.3.1 (GraphPad), nonlinear regression algorithm was used with a constrain of 0 for the bottom and 100 for the top. DNA EXTRACTION AND WHOLE-EXOME SEQUENCING DNA from the tumor,
adjacent non-tumoral liver tissue and organoid was extracted using the Qiagen DNeasy Blood & Tissue kit (Qiagen, Cat. No. 69504) following the manufacturer’s instructions. Extracted DNA
was subjected to whole-exome sequencing. The Twist Human Core Exome kit was used for whole exome capture according to the manufacturer’s guidelines. Sequencing was performed on Illumina
NovaSeq 6000 using paired-end 100-bp (mean sequencing depth 135× for HCC, 153× for the organoid and 129× for germline (adjacent non-tumoral liver tissue)). Sequencing was performed by CeGaT
(Tübingen, Germany). Reads obtained were aligned to the reference human genome GRCh38 using Burrows-Wheeler Aligner (BWA, v0.7.12)12. Local realignment, duplicate removal, and base quality
adjustment were performed using the Genome Analysis Toolkit (GATK, v4.1 and Picard (http://broadinstitute.github.io/picard/)). Somatic single nucleotide variants (SNVs) and small insertions
and deletions (indels) were detected using MuTect2 (GATK 4.1.4.1)13 and Strelka (v.2.9.10)14. Only variants detected by both callers were kept. We filtered out SNVs and indels outside of the
target regions (i.e., exons), those with a variant allelic fraction (VAF) of <5 % and/or those supported by <3 reads. We excluded variants for which the tumor VAF was <5 times that
of the paired non-tumor VAF. We further excluded variants identified in at least two of a panel of 123 non-tumor samples, captured and sequenced using the same protocols using the artifact
detection mode of MuTect2 implemented in GATK. All indels were manually inspected using the Integrative Genomics Viewer15. FACETS (v.0.5.14)16 was used to identify allele-specific copy
number alterations (CNAs). Genes with total copy number greater than gene-level median ploidy were considered gains; greater than ploidy + 4, amplifications; less than ploidy, losses; and
total copy number of 0, homozygous deletions. Somatic mutations associated with the loss of the wild-type allele (i.e., loss of heterozygosity [LOH]) were identified as those where the
lesser (minor) copy number state at the locus was 0. For chromosome X, the log ratio relative to ploidy was used to call deletions, loss, gains and amplifications. All mutations on
chromosome X in male patients were considered to be associated with LOH. Comparison of copy number between organoids and tumor were performed at gene level. REPORTING SUMMARY Further
information on research design is available in the Nature Research Reporting Summary linked to this article. RESULTS CASE PRESENTATION A 74-year-old man presented with sudden one-sided loss
of vision in the absence of other neurological symptoms. Diagnostic workup revealed giant cell arteritis (GCA) with involvement of the temporal artery, which was confirmed histologically and
treated with aspirin as well as high-dose steroids. An imaging workup was initiated to investigate whether GCA was a paraneoplastic phenomenon. A computed tomography scan revealed a mass
with maximum diameter of 4 cm in the gastric corpus as well as an intrahepatic mass with a maximum diameter of 4 cm. The intrahepatic mass was hypoechogenic in ultrasound and was highly
suspicious for malignancy in the consecutive magnet resonance imaging (MRI) (Fig. 1a, b). Biopsies of the two lesions revealed two independent malignancies: A gastrointestinal stromal tumor
(GIST) of the stomach and a poorly differentiated hepatocellular carcinoma (HCC). Liver values were within the normal range (aspartate aminotransferase (ASAT) of 17 U/l, alanine
aminotransferase (ALAT) of 17 U/l, gamma-glutamyl transferase (GGT) of 30 U/l, alkaline phosphatase (AP) of 77 U/l). Liver function tests were normal, and the radiological findings were not
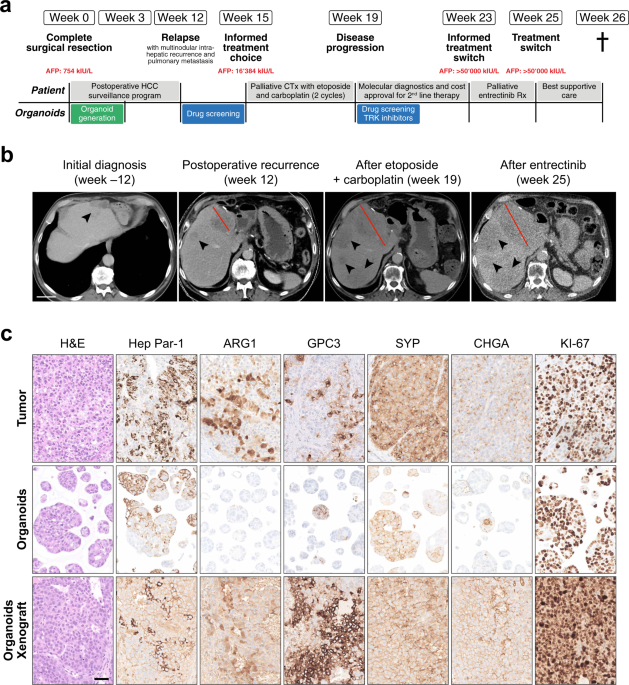
suspicious for advanced liver fibrosis or cirrhosis. Tumor markers revealed high Alpha-Fetoprotein (AFP) levels of 754 kIU/l (normal range: <5.8 kIU/l), as well as slightly elevated
carcinoembryonic antigen (CEA) (4.7 μg/l, normal range: <3.4 μg/l) and carboanhydrate-antigen 19-9 (CA 19-9) (47.7 U/ml, normal range: <34 U/ml) (Fig. 1a). The interdisciplinary tumor
board recommended a curative treatment approach with surgical resection. The patient underwent left-lateral liver resection and simultaneous partial gastrectomy without complications,
allowing complete local resection of both tumors. Histopathological assessment confirmed the diagnosis of GIST of the stomach (Supplementary Fig. 1a). Based on the tumor size of 3.7 cm,
mitotic activity and tumor localization, the GIST had a low risk for progression and required no further therapy17. Macroscopic evaluation of the resected liver tumor showed a polynodular
tumor with beige and partly yellow cut-surface. Microscopy displayed a poorly differentiated tumor composed of medium to large cells with moderate to marked pleomorphism, growing in solid
patternless sheets, lack of sinusoidal spaces and gland formation, compatible with Edmondson–Steiner grade IV HCC10 (Supplementary Fig. 1b). Furthermore, lymphovascular and perineural
invasion could be observed and the tumor was necrotic in about 20% (Supplementary Fig. 2). Immunophenotypic characterization resulted in the definitive diagnosis of poorly differentiated
hepatocellular carcinoma (Edmondson-Steiner grade IV) with neuroendocrine differentiation (HCC-NED). The tumor cells were positive for Hep Par-1, Arginase 1 (ARG1), CD10, Glypican-3 (GPC3),
and KRT19 (Fig. 1c, Supplementary Figs. 1 and 2). The same tumor cells were also positive for the neuroendocrine markers Synaptophysin (SYP) and Chromogranin (CHGA) (Fig. 1c and
Supplementary Fig. 2). Weak positive staining for somatostatin-receptor 2 (SSTR2) was detected in 10% of the tumor cells (Supplementary Fig. 1c). In contrast CD56 was negative (Supplementary
Fig. 1c). The proliferation marker KI-67 was expressed in 85% of the tumor cells, documenting a very high proliferation rate (Fig. 1c and Supplementary Fig. 2). The adjacent non-tumoral
liver displayed no substantial alterations (Supplementary Figs. 1b and 2). According to the recommendations of the interdisciplinary tumor board, the patient was enrolled into a
postoperative HCC surveillance program. The first computed tomography (CT) scan three months after surgery revealed multifocal intrahepatic disease recurrence (max. diameter of 8 cm) with
portal vein invasion as well as pulmonary metastasis, requiring palliative systemic therapy (Fig. 1a–c). Because HCC-NEDs are exceedingly rare, there was no published evidence to guide drug
selection. The tumor board tentatively recommended drugs approved for the treatment of HCC but was well aware of the risk that the neuroendocrine differentiation of the tumor might limit the
response to these drugs. PATIENT-DERIVED HCC-NED ORGANOIDS AS A PRECLINICAL TUMOR MODEL Within the last few years, patient-derived organoids have emerged as powerful preclinical model
system to assess drug responsiveness of tumor cells in vitro, thereby allowing personalized medicine18. Accordingly, we generated organoids from the patient’s resected HCC-NED tissue.
HCC-NED organoids grew rapidly after initial seeding, allowing their expansion and characterization within a short time frame of 3 weeks (compared to an average model generation time of 8
weeks for HCC organoids8). Morphologically, HCC-NED organoids presented as solid spheroids comparably to other HCC organoid models8. Histologically, HCC-NED organoids retained growth pattern
and differentiation grade of the original tumor (Fig. 1c). Notably, markers of hepatic (Hep Par-1) and neuroendocrine (SYP, CHGA) differentiation were equally retained in the organoids,
further underlining their ability to recapitulate the tumor biology in vitro. Moreover, to assess their in vivo tumorigenicity, HCC-NED organoids were subcutaneously injected in
immunodeficient mice. HCC-NED organoids could indeed easily be propagated as xenografts. The xenografts retained histological features as well as marker expression reminiscent of the
patient’s primary tumor, including the very high expression rate of the proliferation marker KI-67 (Fig. 1c and Supplementary Fig. 1c). HCC-NED DISPLAYS MUTATIONS IN _TP53_, _CTNNB1_ AND
_NTRK1_ It has been shown that the genetic profile can influence clinical decision-making and treatment selection in several cancer types19. Therefore, we performed whole-exome sequencing
(WES) of the HCC-NED tumor (mean coverage 136×) matched to the non-tumoral liver tissue (mean coverage 130×) and its derived organoids (mean coverage 154×) to identify targetable
alterations. We detected 111 and 116 somatic mutations in the HCC-NED tumor and HCC-NED organoids, respectively (Supplementary Data 1). Of those, 106 were shared between both samples (87.6%;
Fig. 2a). Moreover, analysis of genome-wide copy number alterations detected by WES showed a 71 % correlation between HCC-NED tumor and HCC-NED organoid including the loss of chr 5, 8p, 1q,
gain of chr 13p, 20p and the focal amplification of the 19q12 locus (Supplementary Fig. 3a). The genomic analysis revealed that the HCC-NED tumor harbored _CTNNB1_ (p.S45P) and _TP53_
(p.R273C) hotspot mutations (Fig. 2b). Both genes are frequently mutated in HCC20. TP53 is also commonly mutated in gastroenteropancreatic neuroendocrine cancers21. Furthermore, _NTRK1_,
encoding the Neurotrophic Receptor Tyrosine Kinase 1, was found to harbor a missense variant (p.T741P) of unknown significance (VUS) in the tyrosine kinase (TK) domain in tumor and matched
organoids (Fig. 2b, Supplementary Fig. 3b and Supplementary Data 1). HCC-NED ORGANOIDS ARE SENSITIVE TOWARDS CARBOPLATIN, ETOPOSIDE AND ENTRECTINIB Because HCC with neuroendocrine
differentiation is a very rare entity, treatment allocation is still unclear and relies on documentations found in single case reports. As a first treatment option for this patient, the
combination of atezolizumab and bevacizumab was considered, because it is the current first-line therapy for advanced HCC20,22. However, immunostaining revealed the lack of Human Leucocyte
Antigen (HLA) ABC and Programmed Death-Ligand 1 (PD-L1) expression on tumor cells (Supplementary Fig. 1c, e), and therefore, the efficacy of immune checkpoint inhibitor therapy might be
impaired in this patient23,24,25. Current guidelines recommend a limited number of systemic treatments for advanced HCC or for NET/NEC21,26 (Supplementary Fig. 4a). To identify potentially
effective drugs, we used the HCC-NED organoids for in vitro drug response testing. 4 HCC organoids from our biobank (Supplementary Data 2) served as controls. Drug screenings were performed
with a broad drug dilution range of 0.02 nM to 10 μM. Cells were treated for six days at which time the cell number was then determined using an ATP-based readout as described in the
materials and methods. HCC-NED organoids showed the same response to the multikinase-inhibitors sorafenib, lenvatinib, cabozantinib and regorafenib as the control HCC organoid lines (Fig.
2c, Supplementary Fig. 4b, Supplementary Data 3 and 4). We then tested drugs approved for advanced neuroendocrine tumors and carcinomas, including somatostatin analogs (octreotide,
lanreotide, pasireotide), the multikinase-inhibitor sunitinib, the mTOR-inhibitor everolimus as well as conventional chemotherapeutics (5-FU, cisplatin, etoposide, carboplatin). Compared to
conventional HCC organoids, HCC-NED organoids responded better to the classical chemotherapeutics cisplatin, etoposide and carboplatin (Fig. 2c, d, Supplementary Fig. 4c, Supplementary Data
3 and 4). Of note, no antitumoral activity could be observed for any of the somatostatin analogs. We also tested the pan-TRK inhibitors entrectinib and larotrectinib. In a recent report,
these drugs were strong growth inhibitors of organoids derived from gastroenteropancreatic neuroendocrine neoplasms27. Larotrectinib had no effect on our HCC-NED and HCC organoids (Fig. 2c,
Supplementary Fig. 4d and Supplementary Data 4). On the other hand, HCC-NED organoids were growth inhibited by entrectinib with an IC50 of 0.61 μM (Fig. 2c, d, Supplementary Data 3 and 4).
PROGRESSIVE DISEASE AFTER TWO CYCLES OF ETOPOSIDE AND CARBOPLATIN, FOLLOWED BY A TREATMENT ATTEMPT WITH ENTRECTINIB Based on the above-mentioned results, a first-line palliative chemotherapy
with etoposide and carboplatin was initiated. The therapy was well tolerated by the patient. However, a CT scan after 4 weeks revealed progressive disease (Fig. 1b), and the treatment was
stopped after only two cycles. Because next-generation sequencing analysis of the tumor had revealed a _NTRK1_-mutation and the pan-TRK inhibitor entrectinib was effective in HCC-NED
organoids, a therapy attempt with entrectinib was suggested for second line. After patient consultation, interdisciplinary discussion at the tumor board, and approval by the health insurance
company, entrectinib treatment was initiated 4 weeks later. Unfortunately, the patient’s general health condition had rapidly deteriorated and he additionally suffered a traumatic femoral
neck fracture. Entrectinib treatment was still initiated but had to be discontinued after two weeks. A CT scan revealed progression of the number and size of the metastatic lesions, but also
increasing areas without up-take of contrast material (non-viable tumor) (Fig. 1b). A formal evaluation of the response to entrectinib was not possible due to the short treatment period.
The patient then received best supportive care. He deceased one week later. DISCUSSION Hepatocellular carcinoma with neuroendocrine differentiation is a very rare tumor entity1,2. No
evidence-based treatment options are established for these aggressive primary liver carcinomas. Published reports describe the use of surgical resection, percutaneous ablation, transarterial
chemoembolization and classical systemic chemotherapies4. However, most reports describe a poor outcome despite these treatments. In the present report, we describe for the first time a
functional precision oncology approach to guide the choice of systemic chemotherapeutics in liver cancer. We successfully generated tumor organoids that were then used for testing the
efficacy of drugs in vitro. The HCC-NED organoid line grew very rapidly, probably reflecting the exceedingly high proliferation rate of the originating tumor. It is known that high
proliferation rates increase the success rate for generating organoids from tumor biopsies8. We therefore believe that HCC-NECs and HCC-NEDs are good candidates for generating tumor
organoids. Furthermore, the rapid growth of HCC-NED organoids allowed their characterization as well as drug testing within a time frame of 5–6 weeks, the latter being an important factor
when using pre-clinical models for therapy guidance. Indeed, the applicability of tumor organoid models in the clinical setting strongly depends on the time scale of establishment28,29,30.
In our case, despite good efficacy in the organoid tumor model in vitro, the combination of etoposide and carboplatin as first-line palliative therapy was clinically not effective. We can
only speculate about the reasons for this failure. Clearly, the organoid models, while maintaining most of the key cellular and molecular features of the originating tumors, have important
limitations because they lack the tumor stroma. This precludes testing anti-angiogenic drugs such as ramucirumab or immune-checkpoint inhibitors. It is also conceivable that the tumor stroma
influences the response to treatments targeted to the tumor cells. Such effects cannot be assessed in the organoid models. It is also possible that the drug concentrations in the tumor were
too low, or that the tumors developed rapid resistance to etoposide and carboplatin in vivo. Fusions involving _NTRK_ genes are the most common mechanisms of oncogenic TRK activation31.
Typically, the fusions contain 3’ sequences of _NTRK_s that include the kinase domain and 5’ sequences of a different gene. The fusion results in a chimeric oncoprotein with
ligand-independent constitutive activation of the TRK kinase32. Clinical detection of _NTRK_ fusions is mainly based on next-generation sequencing (NGS). Immunohistochemistry is a
complementary method that can detect TRK overexpression as a surrogate for _NTRK_ fusions33,34. The missense mutation present in our case (T741P) was not associated with TRK overexpression,
since both HCC-NED tissue and organoids stained negative in pan-TRK IHC (Supplementary Fig. 1d). An increasing number of _NTRK_ mutations and splice variants and cases of TRK ectopic
expression and/or overexpression has been reported32. However, overall _NTRK1_ is not frequently mutated in neuroendocrine tumors such as pancreas35,36 and prostate37 (0 and 1.2% frequency,
respectively), in HCC the frequency is 0.7%38,39,40,41,42,43. For most of these _NTRK_ mutations, the functional consequences and their role as oncogenic drivers are unknown or remain
controversial. In our case, we have no evidence that the mutation in the kinase domain is indeed a driver mutation. The observation that HCC-NED organoids did not respond to the selective
pan-TRK inhibitor larotrectinib, but were sensitive to entrectinib, a pan-TRK inhibitor with additional activity against the proto-oncogene kinase ROS1 and anaplastic lymphoma kinase (ALK)44
does not support the hypothesis that constitutive TRK activity was a main oncogenic driver in our case. Moreover, _NTRK1_ (T741P) was predicted to be deleterious by the MetaSV score45. Of
note, this is only based on in silico predictions and further studies are required to unveil the functional impact of this mutation. Limited resources did not allow us to test all
conventional chemotherapeutics listed in the guidelines for NET/NEC treatment21. Specifically, we did not investigate the antitumoral efficacy of temzolomide, streptozocin, capecitabine,
leucovorin, oxaliplatin and irinotecan in our HCC-NED organoid model. Accordingly, we cannot rule out that these cytostatic drugs might have displayed antitumoral activity. The efficacy of
these components can be tested to potentially inform future treatment decisions. Furthermore, this unique HCC-NED organoid line can be used to screen additional drug libraries, an effort
that might identify promising candidates for future cases of HCC-NED. In conclusion, we describe a rare case of a patient with HCC with neuroendocrine differentiation. The rapid
establishment of patient-derived tumor organoids allowed in vitro drug testing and thereby personalized treatment choices in this patient. Unfortunately, the drugs could not prevent the
rapid tumor progression. Nevertheless, the report provides a first proof-of-principle for using organoids for personalized medicine in these rare primary liver cancers. DATA AVAILABILITY The
WES data reported here are available under restricted access at the European Genome- Phenome Archive under primary accession number EGAS00001005887. Access is restricted because genetic
data is personally identifiable. To obtain access and conditions of access to the EGA datasets, contact the corresponding authors, who will respond within 4 weeks. The use of the data will
be subjected to agreement of a data use policy, which details the minimum protection measures required related to data encryption and user access. The data will be available to the
authorized users for the duration of the requested project. Users will have to specifically agree to preserve, at all times, the confidentiality of information and Data pertaining to Data
Subjects and to use or attempt to use the Data to compromise or otherwise infringe the confidentiality of information on Data Subjects and their right to privacy. User have to agree not to
attempt to identify Data Subjects. The full data use policy will be available upon data access request. Source data for Fig. 2 panels a and b can be found in Supplementary Data 1. Source
data for Fig. 2 panel c can be found in Supplementary Data 3. Source data for Fig. 2 panel d, and Supplementary Fig. 4 panels b, c, and d, can be found in Supplementary Data 4. REFERENCES *
Torbenson, M. S. Morphologic subtypes of hepatocellular carcinoma. _Gastroenterol. Clin. North Am._ 46, 365–391 (2017). Article Google Scholar * Nomura, Y. et al. Clinicopathological
features of neoplasms with neuroendocrine differentiation occurring in the liver. _J. Clin. Pathol._ 70, 563–570 (2017). Article CAS Google Scholar * La Rosa, S., Sessa, F. & Uccella,
S. Mixed neuroendocrine-nonneuroendocrine neoplasms (MiNENs): unifying the concept of a heterogeneous group of neoplasms. _Endocr. Pathol._ 27, 284–311 (2016). Article Google Scholar *
Nakano, A. et al. Combined primary hepatic neuroendocrine carcinoma and hepatocellular carcinoma: case report and literature review. _World J. Surg. Oncol._ 19, 78 (2021). Article Google
Scholar * Lu, J. G., Farukhi, M. A., Mayeda, D. & French, S. W. Hepatocellular carcinoma with neuroendocrine differentiation: a case report. _Exp. Mol. Pathol._ 103, 200–203 (2017).
Article CAS Google Scholar * Jahan, N., Warraich, I., Onkendi, E. & Awasthi, S. Mixed hepatocellular carcinoma-neuroendocrine carcinoma—a diagnostic and therapeutic challenge.
_Current Probl. Cancer Case Rep._ 1, 100020 (2020). * Okumura, Y. et al. Combined primary hepatic neuroendocrine carcinoma and hepatocellular carcinoma with aggressive biological behavior
(adverse clinical course): a case report. _Pathol. Res. Pract._ 213, 1322–1326 (2017). Article Google Scholar * Nuciforo, S. et al. Organoid models of human liver cancers derived from
tumor needle biopsies. _Cell Rep._ 24, 1363–1376 (2018). Article CAS Google Scholar * Broutier, L. et al. Human primary liver cancer-derived organoid cultures for disease modeling and
drug screening. _Nat. Med._ 23, 1424–1435 (2017). Article CAS Google Scholar * Edmondson, H. A. & Steiner, P. E. Primary carcinoma of the liver: a study of 100 cases among 48,900
necropsies. _Cancer_ 7, 462–503 (1954). Article CAS Google Scholar * Torbenson, M. S. et al. Hepatocellular carcinoma. In: _WHO classification of tumours: digestive system tumours_, 5th
ed. (International Agency for Research on Cancer, 2019). * Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. _Bioinformatics_ 26, 589–595 (2010).
Article Google Scholar * Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. _Nat. Biotechnol._ 31, 213–219 (2013). Article CAS
Google Scholar * Saunders, C. T. et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. _Bioinformatics_ 28, 1811–1817 (2012). Article CAS
Google Scholar * Thorvaldsdottir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. _Brief
Bioinform._ 14, 178–192 (2013). Article CAS Google Scholar * Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput
DNA sequencing. _Nucleic Acids Res._ 44, e131 (2016). Article Google Scholar * Miettinen, M. & Lasota, J. Gastrointestinal stromal tumors: pathology and prognosis at different sites.
_Semin. Diagn. Pathol._ 23, 70–83 (2006). Article Google Scholar * Nuciforo, S. & Heim, M. H. Organoids to model liver disease. _JHEP Rep._ 3, 100198 (2021). Article Google Scholar *
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. _Nat. Med._ 23, 703–713 (2017). Article CAS Google Scholar *
Llovet, J. M. et al. Hepatocellular carcinoma. _Nat. Rev. Dis. Primers_ 7, 6 (2021). Article Google Scholar * Pavel, M. et al. Gastroenteropancreatic neuroendocrine neoplasms: ESMO
Clinical Practice Guidelines for diagnosis, treatment and follow-up. _Ann. Oncol._ 31, 844–860 (2020). Article CAS Google Scholar * Finn, R. S. et al. Atezolizumab plus bevacizumab in
unresectable hepatocellular carcinoma. _N. Engl. J. Med._ 382, 1894–1905 (2020). Article CAS Google Scholar * Rodig, S. J. et al. MHC proteins confer differential sensitivity to CTLA-4
and PD-1 blockade in untreated metastatic melanoma. _Sci. Transl. Med._ 10, eaar3342 (2018). * Lee, J. H. et al. Transcriptional downregulation of MHC class I and melanoma de-
differentiation in resistance to PD-1 inhibition. _Nat. Commun._ 11, 1897 (2020). Article CAS Google Scholar * Sangro, B. et al. Association of inflammatory biomarkers with clinical
outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. _J. Hepatol._ 73, 1460–1469 (2020). Article CAS Google Scholar * European Association for the Study of the
Liver. Electronic address: [email protected]; European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. _J. Hepatol._
69, 182–236 (2018). * Kawasaki, K. et al. An organoid biobank of neuroendocrine neoplasms enables genotype-phenotype mapping. _Cell_ 183, 1420–1435.e1421 (2020). Article CAS Google
Scholar * Pauli, C., et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. _Cancer Discov_ (2017). * Bose, S., Clevers, H. & Shen, X. Promises and
challenges of organoid-guided precision medicine. _Medicine_ 2, 1011–1026 (2021). Article Google Scholar * Wensink, G. E. et al. Patient-derived organoids as a predictive biomarker for
treatment response in cancer patients. _NPJ Precis. Oncol._ 5, 30 (2021). Article Google Scholar * Vaishnavi, A. et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer.
_Nat. Med._ 19, 1469–1472 (2013). Article CAS Google Scholar * Cocco, E., Scaltriti, M. & Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. _Nat. Rev. Clin. Oncol._
15, 731–747 (2018). Article CAS Google Scholar * Hechtman, J. F. et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. _Am. J. Surg.
Pathol._ 41, 1547–1551 (2017). Article Google Scholar * Rudzinski, E. R. et al. Pan-Trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. _Am. J. Surg.
Pathol._ 42, 927–935 (2018). Article Google Scholar * Scarpa, A. et al. Whole-genome landscape of pancreatic neuroendocrine tumours. _Nature_ 543, 65–71 (2017). Article CAS Google
Scholar * Jiao, Y. et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. _Science_ 331, 1199–1203 (2011). Article CAS Google Scholar
* Beltran, H. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. _Nat. Med._ 22, 298–305 (2016). Article CAS Google Scholar * Pilati, C. et al.
Genomic profiling of hepatocellular adenomas reveals recurrent FRK-activating mutations and the mechanisms of malignant transformation. _Cancer Cell_ 25, 428–441 (2014). Article CAS Google
Scholar * Harding, J. J. et al. Prospective genotyping of hepatocellular carcinoma: clinical implications of next-generation sequencing for matching patients to targeted and immune
therapies. _Clin. Cancer Res._ 25, 2116–2126 (2019). Article CAS Google Scholar * Schulze, K. et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and
potential therapeutic targets. _Nat. Genet._ 47, 505–511 (2015). Article CAS Google Scholar * Ahn, S. M. et al. Genomic portrait of resectable hepatocellular carcinomas: implications of
RB1 and FGF19 aberrations for patient stratification. _Hepatology_ 60, 1972–1982 (2014). Article CAS Google Scholar * Fujimoto, A. et al. Whole-genome sequencing of liver cancers
identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. _Nat. Genet._ 44, 760–764 (2012). Article CAS Google Scholar * Cancer Genome Atlas
Research Network. Electronic address: [email protected]; Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. _Cell_ 169,
1327–1341.e1323 (2017). * Menichincheri, M. et al. Discovery of entrectinib: a new 3-aminoindazole as a potent anaplastic lymphoma kinase (ALK), c-ros oncogene 1 kinase (ROS1), and
pan-tropomyosin receptor kinases (Pan-TRKs) inhibitor. _J. Med. Chem._ 59, 3392–3408 (2016). Article CAS Google Scholar * Kim, S., Jhong, J. H., Lee, J. & Koo, J. Y. Meta-analytic
support vector machine for integrating multiple omics data. _BioData Min._ 10, 2 (2017). Article Google Scholar Download references ACKNOWLEDGEMENTS We thank the patient for study
participation, the operating room caretakers for the support in collecting biological material and Prof. Dr. Katharina Rentsch for serum AFP measurement. We thank Dr. Xueya Wang for the
support with animal experiments and Petra Hirschmann for performing immunohistochemical stainings. Furthermore, we are very grateful for the support of Dr. Diego Calabrese and his team at
the Histology Core Facility of the Department of Biomedicine at the University of Basel. Entrectinib and larotrectinib were a kind gift of Prof. Dr. Alfred Zippelius. This work was funded by
European Research Council Synergy grant 609883 (MERiC) and by SystemsX.ch grant MERiC to M.H.H. S.P. was supported by the Swiss Cancer League (KFS-4988-02-2020-R), by the Theron Foundation,
Vaduz (LI), by the Surgery Department of the University Hospital Basel and by the Prof. Max Clöetta Stiftung. M.A.M. was supported by a personal grant (MD-PhD-4500-06-2018) of the Swiss
Cancer Research Foundation. L.M.T. was supported by AIRC grant number IG 2019 Id.23615. The funding bodies had no role in study design; in the collection, analysis and interpretation of
data; in the writing of the report; and in the decision to submit the article for publication. AUTHOR INFORMATION Author notes * These authors contributed equally: Marie-Anne Meier, Sandro
Nuciforo, Mairene Coto-Llerena. AUTHORS AND AFFILIATIONS * Department of Biomedicine, University Hospital and University of Basel, CH-4031, Basel, Switzerland Marie-Anne Meier, Sandro
Nuciforo, Mairene Coto-Llerena, John Gallon, Salvatore Piscuoglio & Markus H. Heim * Clarunis University Center for Gastrointestinal and Liver Diseases, CH-4002, Basel, Switzerland
Marie-Anne Meier, Savas D. Soysal, Otto Kollmar & Markus H. Heim * Institute of Medical Genetics and Pathology, University Hospital Basel, CH-4031, Basel, Switzerland Mairene
Coto-Llerena, Matthias S. Matter, Caner Ercan, Jürg Vosbeck & Salvatore Piscuoglio * Department of Anatomic Pathology, IRCCS Humanitas Research Hospital, Rozzano, Milan, Italy Luigi M.
Terracciano * Humanitas University, Department of Biomedical Sciences, Pieve Emanuele, Milan, Italy Luigi M. Terracciano * Radiology and Nuclear Medicine, University Hospital Basel, CH-4031,
Basel, Switzerland Daniel Boll * Department of Oncology, University Hospital Basel, CH-4031, Basel, Switzerland Raphaël Delaloye Authors * Marie-Anne Meier View author publications You can
also search for this author inPubMed Google Scholar * Sandro Nuciforo View author publications You can also search for this author inPubMed Google Scholar * Mairene Coto-Llerena View author
publications You can also search for this author inPubMed Google Scholar * John Gallon View author publications You can also search for this author inPubMed Google Scholar * Matthias S.
Matter View author publications You can also search for this author inPubMed Google Scholar * Caner Ercan View author publications You can also search for this author inPubMed Google Scholar
* Jürg Vosbeck View author publications You can also search for this author inPubMed Google Scholar * Luigi M. Terracciano View author publications You can also search for this author
inPubMed Google Scholar * Savas D. Soysal View author publications You can also search for this author inPubMed Google Scholar * Daniel Boll View author publications You can also search for
this author inPubMed Google Scholar * Otto Kollmar View author publications You can also search for this author inPubMed Google Scholar * Raphaël Delaloye View author publications You can
also search for this author inPubMed Google Scholar * Salvatore Piscuoglio View author publications You can also search for this author inPubMed Google Scholar * Markus H. Heim View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.A.M., S.N., M.C.L., S.P. and M.H.H. conceived the study and designed the experiments; R.D., S.D.S.,
O.K., D.B. and M.H.H. recruited the patient and were involved in patient care; M.S.M., C.E., J.V. and L.M.T. performed histopathological analyses of the resected tissue; M.A.M., S.N.,
M.S.M., M.C.L. and J.G. performed experiments and analyzed the data; M.A.M., S.N., M.C.L., M.S.M., S.P. and M.H.H. wrote the manuscript; M.H.H. and S.P. obtained funding. CORRESPONDING
AUTHORS Correspondence to Salvatore Piscuoglio or Markus H. Heim. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interest. PEER REVIEW PEER REVIEW INFORMATION
_Communications Medicine_ thanks Meritxell Huch, Jörg Trojan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are
available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY INFORMATION SUPPLEMENTARY DATA 1 SUPPLEMENTARY DATA 2 SUPPLEMENTARY DATA 3 SUPPLEMENTARY DATA 4 PEER REVIEW FILE
REPORTING SUMMARY RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation,
distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and
indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to
the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will
need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Meier, MA., Nuciforo, S., Coto-Llerena, M. _et al._ Patient-derived tumor organoids for personalized medicine in a patient with rare hepatocellular carcinoma with
neuroendocrine differentiation: a case report. _Commun Med_ 2, 80 (2022). https://doi.org/10.1038/s43856-022-00150-3 Download citation * Received: 21 September 2021 * Accepted: 21 June 2022
* Published: 01 July 2022 * DOI: https://doi.org/10.1038/s43856-022-00150-3 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link
Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative