Association of lncrna-pax8-as1 and lair-2 polymorphisms along with their expression with clinical and subclinical hypothyroidism
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The genetic and epigenetic architecture of clinical and subclinical hypothyroidism remains unclear. We investigated the impact of _long noncoding RNA (LncRNA)-PAX8-AS1_ and _LAIR-2_
genetic variants on the susceptibility to clinical and subclinical hypothyroidism, their influence on LncRNA-PAX8-AS1 and LAIR-2 expression and their potential as hypothyroid biomarkers.
Hundred clinical hypothyroid patients, 110 subclinical hypothyroid patients, and 95 healthy controls were enrolled. Gene expression analysis and genotyping were performed by qPCR. LAIR-2
protein, a proinflammatory mediator, was tested by ELISA. Serum LncRNA-PAX8-AS1 was downregulated, whereas LAIR-2 mRNA and protein levels were upregulated in clinical and subclinical
hypothyroid patients compared to healthy controls. _LncRNA-PAX8-AS1_ rs4848320 and rs1110839 were associated with increased risk of clinical hypothyroidism. Interestingly, both SNPs were
associated with differential expression of serum LncRNA-PAX8-AS1 among clinical hypothyroid patients_. LAIR-2_ rs2287828 was associated with elevated risk of both clinical and subclinical
hypothyroidism. Harboring the rs2287828 T allele augmented the LAIR-2 mRNA expression among clinical hypothyroid patients, while elevated both LAIR-2 mRNA and protein levels in subclinical
hypothyroid patients. The rs4848320-rs1110839-rs2287828 TTT, CTT, and CGT haplotypes were associated with increased hypothyroid risk. Surprisingly, serum LncRNA-PAX8-AS1 and LAIR-2 mRNA
expression demonstrated superior diagnostic accuracy for clinical hypothyroidism and turned out as independent predictors in the multivariate analysis. Conclusively, _LncRNA-PAX8-AS1_ and
_LAIR-2_ genetic variants are novel genetic biomarkers of hypothyroidism that could alter the LncRNA-PAX8-AS1 and LAIR-2 expression. LncRNA-PAX8-AS1 and LAIR-2 expression profiles have the
potential as effective diagnostic and prognostic indicators of hypothyroidism. SIMILAR CONTENT BEING VIEWED BY OTHERS CHARACTERIZING THE FUNCTION OF _EPB41L4A_ IN THE PREDISPOSITION TO
PAPILLARY THYROID CARCINOMA Article Open access 17 November 2020 RS4911154 OF CIRC-ITCH AGGRAVATED TUMOR MALIGNANCY OF THYROID NODULES VIA THE CIRC-ITCH/MIR-22-3P/CBL AXIS Article Open
access 16 September 2021 MUTATED LNCRNA INCREASE THE RISK OF TYPE 2 DIABETES BY PROMOTING Β CELL DYSFUNCTION AND INSULIN RESISTANCE Article Open access 27 October 2022 INTRODUCTION
Hypothyroidism is a common condition with devastating health consequences that affect about 5% of the general population. The declined thyroid hormones secretion evokes a decrease in the
metabolic rate and can incur a life-threatening myxedema and other symptoms resulting in a great social and economic encumbrance1,2. According to the degree of thyroid function reduction,
clinical and subclinical hypothyroidism cases were distinguished1,2. Subclinical hypothyroidism, the earliest biochemical abnormality in hypothyroidism, is characterized by mildly elevated
serum thyrotropin or thyroid stimulating hormone (TSH) levels recording > 4.5 mIU/L but < 10 mIU/L, associated with normal serum thyroxine (T4) and triiodothyronine (T3) concentrations
in the absence of obvious clinical manifestations. While clinical hypothyroidism, the latest biochemical abnormality in hypothyroidism, is recognized by higher levels of TSH (> 10
mIU/L), associated with a decline in both serum T3 and T4 levels, at which stage most patients have obvious clinical symptoms (overt hypothyroidism)3,4. Interestingly, the prevalence of
subclinical hypothyroidism is substantially higher than that of overt hypothyroidism (4 to 10% versus 0.1 to 2%, respectively) in the general population. When non-age-specific TSH reference
values are employed, up to 15% of persons 65 years or older have subclinical hypothyroidism. Women are nearly ten times more likely than men to develop spontaneous hypothyroidism4,5. Despite
such high incidence, the pathogenesis of subclinical hypothyroidism remains elusive. Thereby, decoding the molecular underpinnings of thyroid pathology could identify new potential targets
for early detection and treatment of subclinical hypothyroidism, and hence slowing down the progression to clinical hypothyroidism. Besides the well-known causative factors of
hypothyroidism–iodine deficiency and autoimmunity5-genetic, epigenetic, and environmental factors are implicated in its development by altering the thyroid function6,7; however, studies on
such genetic and epigenetic molecules are still lacking. Long noncoding RNAs (lncRNAs, longer than 200 nucleotides in length) fine-tune gene expression by recruiting epigenetic complexes or
directly affecting transcription of multiple genes acting on various cellular paradigms8,9. The expression of lncRNAs is exceedingly controlled, with organ, tissue and even cellular specific
patterns8,10 and they have been documented to have crucial roles in the normal development, function, physiology, and pathophysiology of several endocrine organs such as the prostate,
pancreas, and adipose tissue11,12. However, little is known about the biological function of lncRNAs and their expression pattern in the thyroid gland. Importantly, several molecules were
deduced to function in hypothyroidism, the lncRNA-paired-box gene 8-antisense RNA 1 (LncRNA-PAX8-AS1) and the leukocyte-associated Ig-like receptor-2 (LAIR-2) were proposed as one of the
most important in such condition1,13. The _LncRNA-PAX8-AS1_, mapped to chromosome 2q13 upstream of _PAX8_ gene, is a potential regulator of PAX814. PAX8 was regarded as a nuclear
transcription factor of the mammalian PAX protein family in the developing and adult thyroid15,16. Interestingly, PAX8 is the master regulator of the differentiated thyroid phenotype and is
required in thyroid cells for transcriptional activation of thyroglobulin, thyroid peroxidase, and sodium iodide symporter16. An expression quantitative trait locus (eQTL) is a locus
containing a genetic variant that impact the expression level of a gene17,18. The _LncRNA-PAX8-AS1_ gene locus has been shown to encompass polymorphisms that represent eQTLs for PAX8.
Specifically, the single nucleotide polymorphisms (SNPs) rs4848320 and rs1110839 in the _LncRNA-PAX8-AS1_ gene are candidate eQTLs of PAX8 as revealed by previous bioinformatics analyses.
These studies raised the possibility that these variants might influence the expression, function or structure of the LncRNA-PAX8-AS1, thereby amending the expression of PAX814,19. However,
the mechanistic role of these SNPs and whether they govern the expression of LncRNA-PAX8-AS1 in hypothyroidism are not previously explored. The leukocyte-associated Ig-like receptor (LAIR)
is a small family of immune receptor tyrosine-based inhibitory motifs (ITIM)-containing inhibitory receptor that belongs to the Ig superfamily20. _LAIR-2_ gene is located on human chromosome
19q13.4. It encodes LAIR-2 protein, a secreted receptor acting as a pro-inflammatory mediator through limiting the potential of the immune inhibitor LAIR-1_._ This results in enhanced
activation of immune cells, the hallmark of autoimmune hypothyroid disease13,21. Importantly, LAIR molecules interact with collagens with high affinity. Peculiarly, the interaction of LAIR-1
with collagens directly inhibited immunocyte activation in vitro, suggesting a vital mechanism of peripheral immune regulation through the extracellular matrix21. LAIR-2 plays a significant
role in such regulation based on its higher affinity for collagens than LAIR-1. Indeed, elevated levels of serum LAIR-2 exhibited a potential regulatory capability in modulating the
inhibitory power of LAIR-1 in patients with autoimmune thyroid diseases13. It has been found that some SNPs located near the _LAIR-2_ gene may influence the susceptibility to autoimmune
diseases. For instance, the _LAIR-2_ SNP rs2287828 has been shown to be associated with the susceptibility to ankylosing spondylitis and pemphigus diseases and was found to affect the LAIR-2
mRNA expression level22,23. However, the impact of this SNP on the risk of hypothyroid disease and its subclinical condition as well as its possible control on LAIR-2 mRNA and protein
expression in hypothyroidism remains to be investigated. Therefore, this study aimed to investigate the possible role of _LncRNA-PAX8-AS1_ and _LAIR-2_ genetic variants in the development of
both subclinical and clinical (overt) hypothyroidism. The study also explored the expression pattern of LncRNA-PAX8-AS1, LAIR-2 mRNA and protein, their diagnostic and prognostic potential,
and their correlations with clinical data in both conditions. The mechanistic influence of the studied SNPs on gene expression and their association with patients’ parameters were also
assessed. RESULTS DEMOGRAPHIC AND CLINICAL CHARACTERISTICS OF THE STUDIED GROUPS As shown in Table 1, both hypothyroid groups have the same age and gender distribution. Comparing with
healthy controls, hypothyroid patients exhibited significantly higher body mass index (BMI), thyroid volume, serum TSH, total cholesterol, LDL-cholesterol, triglycerides, fasting plasma
glucose, 2-h postprandial (2HPP) plasma glucose, blood glycated hemoglobin (HbA1c) and fasting plasma insulin, along with significantly lower serum HDL-cholesterol levels (_P_ < 0.05).
However, clinical hypothyroid patients exhibited significantly higher thyroid volume, serum TSH, triglycerides, fasting plasma glucose, 2HPP blood glucose, HbA1c and fasting plasma insulin
levels when compared with levels in the subclinical hypothyroid patients (_P_ < 0.05). On the other hand, only clinical hypothyroid patients exhibited significantly lower serum levels of
free T4 and free T3 than those in healthy controls (_P_ < 0.05), whereas subclinical hypothyroid patients did not show any significant difference regarding free T4 and T3 levels when
compared with the healthy controls levels (_P_ > 0.05). ASSOCIATION OF RS4848320 (C/T), RS1110839 (G/T) AND RS2287828 (C/T) WITH THE RISK OF CLINICAL AND SUBCLINICAL HYPOTHYROIDISM For
all studied SNPs, the minor allele frequency (MAF) in the control group was > 0.05 (Table 2). In the study controls, MAF was slightly higher than the reported global MAF (T = 0.21) for
rs4848320 and slightly lower than the global MAF (T = 0.48 and T = 0.11) for rs1110839 and rs2287828, respectively (Table 2). However, it was close to that reported for some populations for
the three SNPs as recorded in the Ensembl GRCh38 release 102-November 2020. As displayed in Supplementary Table S1, the distribution of the rs4848320, rs1110839, and rs2287828 genotypes in
the study groups did not deviate from HWE (_P_ > 0.05). The best fit genetic models of rs4848320, rs1110839, and rs2287828 in clinical and subclinical hypothyroid patients against
controls are demonstrated in Table 3. For rs4848320 (C/T) and rs1110839 (G/T) in the _LncRNA-PAX8-AS1_ gene, the minor T allele was associated with increased risk of clinical hypothyroidism
(adjusted OR = 2.01, _P_ = 0.0015 and 2.2, _P_ < 0.0001, respectively) with adjustment for age and sex as confounding factors (Table 2). In clinical hypothyroid patients, the rs4848320
and rs1110839 log-additive models were significant and showed a noticeable clinical hypothyroid risk (adjusted OR = 2.26 and 2.55, respectively, _P_ < 0.0001). In addition, among the
subclinical hypothyroid patients, the genotype distribution and allele frequencies of rs4848320 and rs1110839 did not show a statistically significant difference when compared with those in
the healthy controls (_P_ > 0.05) (Table 3). Interestingly, the minor T allele frequency of _LAIR-2_ rs2287828 (C/T) was associated with 8.50 and 6.64-fold increase in the risk of
clinical and subclinical hypothyroidism, respectively (T vs. C allele, _P_ < 0.0001 for each) as shown in Table 2. Moreover, a markedly high risk for clinical and subclinical
hypothyroidism was also depicted from the rs2287828 dominant (CT + TT vs CC) model (Adjusted OR = 14.51 and 8.7, respectively, _P_ < 0.0001) (Table 3). Like the other two SNPs, all
results were adjusted for age and sex as covariates. STRATIFICATION ANALYSIS The effect of rs4848320 (C/T), rs1110839 (G/T), and rs2287828 (C/T) SNPs on clinical and subclinical hypothyroid
risk were further examined in a stratified risk analysis by gender (Supplementary Table S2). Among female patients, rs4848320 (C/T) and rs1110839 (G/T) log additive models revealed high risk
for clinical hypothyroidism (adjusted OR = 2.94 and 2.51, respectively, _P_ < 0.0001). Moreover, rs2287828 dominant model (CT + TT vs CC) showed a markedly elevated susceptibility to
clinical and subclinical hypothyroidism among the female patients (adjusted OR = 21.61 and 12.77, respectively, _P_ < 0.0001). Noticeably, stratified analysis among the male patients did
not show any susceptibility risk (_P_ > 0.05). HAPLOTYPE ANALYSIS The combined impact of the studied gene polymorphisms in all hypothyroid patients compared with healthy controls was
evaluated (Table 4). Results revealed that the rs4848320-rs1110839-rs2287828 TTT, CTT, and CGT haplotypes were associated with increased risk of hypothyroidism by 10.15, 27.45 and 9.42-fold,
respectively (TTT vs CGC, _P_ < 0.0001, CTT vs CGC, _P_ = 0.038, and CGT vs CGC, _P_ = 0.027). Other haplotypes were not statistically associated with the risk of hypothyroidism (_P_ ≥
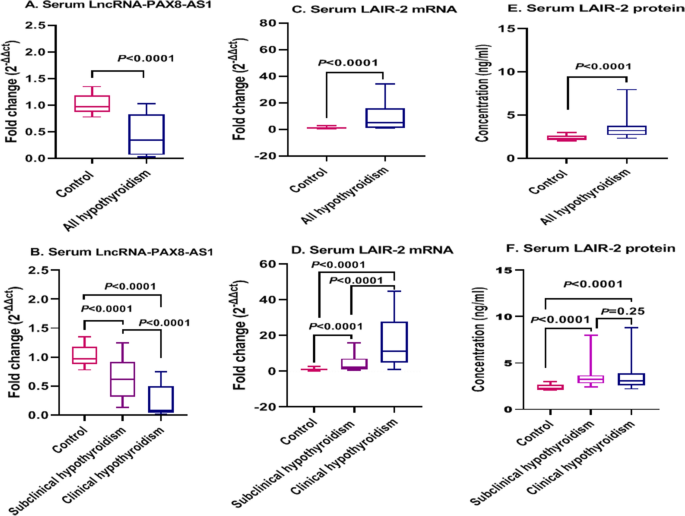
0.05). SERUM LNCRNA-PAX8-AS1 AND LAIR-2 MRNA EXPRESSION LEVELS IN CLINICAL AND SUBCLINICAL HYPOTHYROIDISM As depicted in Fig. 1A, the expression levels of serum lncRNA-PAX8-AS1 were markedly
lower in all hypothyroid groups than those in the healthy controls (_P_ < 0.0001). In-depth analysis showed that serum LncRNA-PAX8-AS1 expression was markedly downregulated with a median
fold change = 0.073 and 0.611 (_P_ < 0.0001 for each) in clinical and subclinical hypothyroid patients, respectively compared with that in the healthy controls, with serum
lncRNA-PAX8-AS1 recorded significantly lower levels in clinical hypothyroid patients than those in the subclinical hypothyroid patients (_P_ < 0.0001) (Fig. 1B). Referring to Fig. 1C,
serum LAIR-2 mRNA expression levels were significantly higher in all hypothyroid groups than those in the healthy controls (_P_ < 0.0001). The detailed results (Fig. 1D) showed that serum
LAIR-2 mRNA expression was upregulated with a median fold change of 11.29 and 2.11 (_P_ < 0.0001 for each) in clinical and subclinical hypothyroid patients, respectively, compared with
that in the healthy controls. Also, clinical hypothyroid patients demonstrated significantly higher LAIR-2 mRNA levels than those in the subclinical hypothyroid patients (_P_ < 0.0001).
SERUM LAIR-2 PROTEIN LEVELS IN CLINICAL AND SUBCLINICAL HYPOTHYROIDISM Serum LAIR-2 protein levels were significantly higher in all hypothyroid groups than the levels in the healthy controls
(_P_ < 0.0001) (Fig. 1E). Interestingly, LAIR-2 protein revealed higher serum levels in clinical and subclinical hypothyroid groups with a median of 3.09 and 3.24 than those in the
healthy controls, respectively (_P_ < 0.0001 for each), nevertheless, there was no significant difference between clinical and subclinical hypothyroid groups (_P_ = 0.25) (Fig. 1F).
ASSOCIATION OF RS4848320, RS1110839, AND RS2287828 SNPS WITH SERUM LNCRNA-PAX8-AS1, LAIR-2 MRNA, AND LAIR-2 PROTEIN LEVELS IN CLINICAL AND SUBCLINICAL HYPOTHYROID PATIENTS Assessment of
serum LncRNA-PAX8-AS1, LAIR-2 mRNA, and LAIR-2 protein levels in clinical and subclinical hypothyroid patients carrying different SNP genotypes was performed in order to study the
mechanistic role of rs4848320, rs1110839, and rs2287828 in hypothyroidism (Fig. 2). For rs4848320, we found that the serum LncRNA-PAX8-AS1 expression levels were significantly higher in
clinical hypothyroid TT genotype carriers than those in the CC genotype carriers (_P_ = 0.0002) (Fig. 2A). Regarding rs1110839, serum LncRNA-PAX8-AS1 expression levels were also markedly
higher in the clinical hypothyroid TT genotype carriers than those in the GG or GT genotype carriers (_P_ < 0.0001) and in the GT genotype carriers than those in the GG carriers (_P_ =
0.0002) (Fig. 2B). Regarding rs2287828, serum LAIR-2 mRNA expression levels were significantly higher in the clinical or subclinical hypothyroid TT genotype carriers than in those carrying
the CC or CT genotype as well as in CT vs CC genotype carriers (_P_ < 0.05). Also, serum LAIR-2 mRNA expression levels were significantly higher in the CT + TT vs CC comparison (_P_ <
0.0001) (Fig. 2C,D). There was no significant difference found in serum LAIR-2 protein levels among the clinical hypothyroid patients carrying different rs2287828 genotypes (_P_ > 0.05)
(Fig. 2E). However, serum LAIR-2 protein levels were significantly higher in the subclinical hypothyroid patients harboring the TT genotype than those having the CC or CT genotypes (_P_ <
0.0001) and in the CT + TT vs CC comparison as well (_P_ = 0.0008) (Fig. 2F). DIAGNOSTIC PERFORMANCE OF LNCRNA-PAX8-AS1, LAIR-2 MRNA, AND LAIR-2 PROTEIN Reciever operating characteristic
(ROC) curve analysis of the studied groups revealed that serum LncRNA-PAX8-AS1 significantly distinguished all hypothyroid group as well as clinical and subclinical hypothyroid patients from
healthy controls with area under the curve (AUC) = 0.88, 0.98, and 0.78, respectively (Fig. 3A–C). In addition, serum LncRNA-PAX8-AS1 discriminated the clinical hypothyroid patients from
subclinical hypothyroid group with AUC = 0.79 (Fig. 3D). These data suggest serum LncRNA-PAX8-AS1 as a potential discriminator for hypothyroidism with an excellent diagnostic performance for
clinical hypothyroid patients (Table 5). Importantly, the analysis also revealed that serum LAIR-2 mRNA distinguished all hypothyroid group, clinical, and subclinical patients (Fig. 3E–G)
from healthy controls with a sufficient performance power (AUC = 0.8, 0.9, and 0.71, respectively) and with an excellent discrimination of clinical hypothyroidism. Moreover, serum LAIR-2
mRNA discriminated the clinical hypothyroid patients from subclinical hypothyroid group with AUC = 0.75 (Fig. 3H). These data highlighted that serum LAIR-2 mRNA expression could be a
promising discriminator of hypothyroidism and its subtypes. LAIR-2 protein discriminated all hypothyroid, clinical, and subclinical hypothyroid patients from healthy controls as well with
AUC = 0.85, 0.81, and 0.89, respectively (Fig. 3I–K), indicating its diagnostic potential. The calculated sensitivities and specificities at the best cutoff values are shown in Table 5.
CORRELATION OF LNCRNA-PAX8-AS1, LAIR-2 MRNA, AND LAIR-2 PROTEIN LEVELS WITH THE DEMOGRAPHIC AND CLINICAL DATA OF CLINICAL HYPOTHYROIDISM As shown in Supplementary Table S3, no significant
correlations were observed for LncRNA-PAX8-AS1 or LAIR-2 mRNA levels with the laboratory data of clinical hypothyroid patients. However, serum LAIR-2 protein levels were positively
correlated with total cholesterol (r = 0.21, _P_ = 0.04) and LDL-cholesterol concentrations (r = 0.28, _P_ = 0.004) in clinical hypothyroid patients. ASSOCIATION OF RS4848320, RS1110839, AND
RS2287828 WITH THE DEMOGRAPHIC AND CLINICAL DATA OF CLINICAL HYPOTHYROIDISM To explore the correlation of rs4848320, rs1110839, and rs2287828 with clinical hypothyroidism, we assessed the
laboratory data in the clinical hypothyroid patients carrying different SNP genotypes (Supplementary Figures S1, S2, and S3). For rs4848320, we found no significant correlations of the
different genotypes with the laboratory data of clinical hypothyroid patients (Supplementary Figure S1). Regarding rs1110839, we found that the clinical hypothyroid patients carrying the GG
genotype had a significant lower serum free T3 levels than those in the TT genotype carriers (_P_ = 0.02) (Supplementary Figure S2-D) and a significant smaller thyroid volume than that in
the GT + TT genotype carriers (_P_ = 0.045) (Supplementary Figure S2-E). In addition, lipid profile assessment in clinical hypothyroid patients carrying different rs1110839 genotypes
revealed higher serum triglyceride levels in the TT genotype carriers than those in the GT (_P_ = 0.017) or GG + GT (_P_ = 0.026) genotype carriers (Supplementary Figure S2-I). However,
sugar profile assessment in clinical hypothyroid patients carrying different rs1110839 genotypes revealed higher fasting plasma glucose levels in those harboring the GG genotype than in
those carrying the GT + TT genotype (_P_ = 0.04) (Supplementary Figure S2-J). Furthermore, the clinical hypothyroid patients carrying the GG genotype exhibited significantly higher 2HPP
plasma glucose, blood HbA1c, and fasting plasma insulin levels than the levels in patients holding the GT or GT + TT genotypes (_P_ < 0.05) (Supplementary Figures S2-K, L, and M,
respectively). Regarding rs2287828, we found that the clinical hypothyroid patients carrying the TT genotype exhibited a significantly higher serum total cholesterol levels than those in the
CC + CT genotypes carriers (_P_ = 0.024) (Supplementary Figure S3-F) and a significantly higher serum triglyceride concentrations than in those harboring the CC or CC + CT genotypes (_P_
< 0.05) (Supplementary Figure S3-I). For the glycemic profile, we found that clinical hypothyroid patients carrying the TT genotype had higher serum fasting plasma insulin levels than
those having the CC + CT genotype carriers (_P_ = 0.047) (Supplementary Figure S3-M). RESULTS OF LOGISTIC REGRESSION ANALYSIS The predictor parameters associated with the risk of clinical
hypothyroidism among both subclinical hypothyroid group and healthy controls were selected using a univariate followed by a multivariate logistic regression analysis (Table 6). Expression
levels of serum LncRNA-PAX8-AS1 and LAIR-2 mRNA were selected as significant predictors associated with the possibilities of clinical hypothyroidism diagnosis in the univariate analysis (_P_
< 0.05). In a stepwise forward multivariate analysis, LncRNA-PAX8-AS1 and LAIR-2 mRNA also turned out to be significant negative and positive predictors of clinical hypothyroid risk,
respectively, in this study (_P_ < 0.05). Additionally, expression levels of LncRNA-PAX8-AS1 and LAIR-2 mRNA as well as its protein turned out as significant final predictors for
subclinical hypothyroid risk among healthy controls in the multivariate analysis (_P_ < 0.05) (Table 7). DISCUSSION Recently, particular attention is paid to discover new biomarkers for
hypothyroidism to reinforce its diagnosis and screening. Furthermore, hormonal replacement over-treatment of hypothyroidism was reported to increase the risk of cardiac morbidity and
mortality, osteoporosis, cognitive dysfunction, and reduced muscle mass; for these reasons novel biomarkers are needed to evolve the therapy3,24,25,26. Dysfunctional regulation of LncRNAs
and mRNAs has been suggested as a novel clue in understanding the pathology of hypothyroidism and thus targeting therapy. However, studies on the impact of _LncRNA-PAX8-AS1_ and _LAIR-2_
gene polymorphisms with differential expression levels on hypothyroidism are still lacking. The current study revealed associations of _LncRNA-PAX8-AS1_ rs4848320 and rs1110839 with
susceptibility to clinical hypothyroidism in our patients, but these SNPs were not associated with subclinical hypothyroidism risk. The study also highlighted the association of _LAIR-2_
rs2287828 with the susceptibility to clinical and subclinical hypothyroidism. Interestingly, the haplotype analysis identified that the joint effect of these three variants in the studied
population was associated with the increased risk of hypothyroidism in all studied patients. This may be interpreted by that patients who have the T allele for the three variants are more
prone to hypothyroidism. Moreover, _LncRNA-PAX8-AS1_ rs4848320 and rs1110839 as well as _LAIR-2_ rs2287828 were functionally correlated with the serum expression levels of LncRNA-PAX8-AS1
and LAIR-2 mRNA along with its protein, respectively in hypothyroidism. These findings may contribute to the pathogenesis of hypothyroidism, and these SNPs could be used as functional
genetic susceptibility markers for sporadic hypothyroidism via functional modulation of LncRNA-PAX8-AS1 and LAIR-2 expression pattern. To our knowledge, this is the first research to
investigate the primary proof of association between _LncRNA-PAX8-AS1_ rs4848320 and rs1110839 as well as _LAIR-2_ rs2287828 with the increased risk of occurrence and progression of
hypothyroidism. The data revealed that the T allele carriers in _LncRNA-PAX8-AS1_ rs4848320 and rs1110839 confer high risk against clinical hypothyroidism, but not subclinical
hypothyroidism. On the other hand, those carrying the T allele in _LAIR-2_ rs2287828 deliberate high risk against both clinical and subclinical hypothyroidism in this study. Surprisingly,
the literature on _LncRNA-PAX8-AS1_ rs4848320 and rs1110839 showed a variety of approaches; in a Southeast Iranian population, the T allele carriers in _LncRNA-PAX8-AS1_ rs4848320 increased
the susceptibility of developing childhood acute lymphocytic leukemia, while rs1110839 variant has no effect19. In the Han Chinese population, T and G allele carriers in _LncRNA-PAX8-AS1_
rs4848320 and rs1110839, respectively, decreased the susceptibility of cervical cancer14. Likewise, the T allele carriers in _LAIR-2_ rs2287828 were associated with the risk of Pemphigus
foliaceus disease in Brazilian population22. However, the mechanistic role of these SNPs was not tackled by these studies. Herein, we found that the studied SNPs affected the differential
gene expression of the circulating LncRNA-PAX8-AS1 and LAIR-2 mRNA coupled with its protein in hypothyroidism. This study identified that serum LncRNA-PAX8-AS1 downregulation as well as
LAIR-2 upregulation could be implicated in hypothyroidism. Also, the multivariate analysis identified serum LncRNA-PAX8-AS1 and LAIR-2 mRNA levels to be independent predictors for being
diagnosed with clinical or subclinical hypothyroidism. Serum LAIR-2 protein was an additional predictor of the diagnosis of subclinical hypothyroidism. Indeed, LncRNA-PAX8-AS1 was
demonstrated as a potential regulator of PAX8, a transcription factor which is crucial for the organogenesis of the developing thyroid gland15. The knockout of _PAX8_ gene has been reported
to reflect the phenotypes of congenital hypothyroid disorders in human patients, delineating disruptions in thyroid hormone biosynthesis (a condition known as dyshormonogenesis)27. Moreover,
in adult thyroid, PAX8 transcription factor was regarded as a master regulator of the differentiated thyroid phenotype by overlapping with genomic binding sites such as promoter regions and
boundary elements close to the 5'-UTRs of identified genes encoding sodium iodide symporter, thyroid peroxidase, and thyroglobulin, thus activating the expression of these critical
proteins for the biosynthesis, storage, and secretion of thyroid hormones; T3 and T415,28. Additionally, the expression level of LncRNA-PAX8-AS1 was previously shown to be significantly
associated with overall or recurrence-free survival time in patients with papillary thyroid carcinoma29. In this study, we identified that downregulation of serum LncRNA-PAX8-AS1 expression
level in clinical and subclinical hypothyroidism may predict the risk and help in the diagnosis of hypothyroidism. The significant lower expression of serum LncRNA-PAX8-AS1 in clinical
hypothyroidism than in subclinical hypothyroidism may foretell the progression risk of hypothyroidism. Importantly, the two SNPs; rs4848320 and rs1110839 in _LncRNA-PAX8-AS1_ gene were
demonstrated as eQTLs of _PAX8_, affecting the expression or function of _LncRNA-PAX8-AS1_, thereby altering the expression of PAX8 that regulates the thyroid gland12,18. Notably, the
finding that _LncRNA-PAX8-AS1_ rs4848320 and rs1110839 were not associated with subclinical hypothyroid patients may justify the normal T3 and T4 levels in these patients. Alternatively, the
association of these SNPs with clinical hypothyroid patients could rationalize in part the lower T3 and T4 in these patients than other groups. These findings reinforce that LncRNA-PAX8-AS1
could be implemented in the occurrence and progression of hypothyroidism. Likewise, LAIR-2 protein, a proinflammatory mediator, that underpins the pathogenesis of autoimmune or inflammatory
diseases through regulating the inhibitory potential of the membrane-bound LAIR-1 by competition for collagen ligands, resulting in enhanced activation of immune cells, a hallmark of
autoimmune hypothyroid diseases20,21. Simone et al.13 documented higher serum levels of LAIR-2 protein in patients with autoimmune thyroid diseases than in healthy controls, possibly due to
different monoclonal antibodies implemented13. The current study showed that LAIR-2 upregulation could predict the risk and help in diagnosis of both clinical and subclinical hypothyroidism.
In clinical hypothyroidism, the expression levels of LncRNA-PAX8-AS1 and LAIR-2 mRNA were markedly different compared to subclinical hypothyroidism and healthy control, indicating their
diagnostic value. However, no significant correlation between LncRNA-PAX8-AS1 and LAIR-2 mRNA with the clinical laboratory data was shown, meanwhile, LAIR-2 protein was positively correlated
with both LDL-cholesterol and total cholesterol levels. Extensive preclinical studies have formerly revealed the critical roles of the innate and adaptive immune systems in promoting
cholesterol-induced atherosclerosis-associated chronic inflammation in arterial blood vessels30. Ample data from previous work31,32,33,34,35,36 revealed that circulating LncRNAs along with
other genes could be promising diagnostic and prognostic biomarkers for different clinical situations. In this study, we displayed that serum LncRNA-PAX8-AS1, LAIR-2 mRNA and its protein
distinguished all hypothyroid patients from healthy controls with moderate to high sensitivity and specificity, and were distinctively expressed between clinical hypothyroid patients and
healthy controls or subclinical hypothyroidism with high accuracy, sensitivity, and specificity, implying that they could be used as potential biomarkers for early and new diagnosis of
hypothyroidism. Moreover, LncRNA-PAX8-AS1 and LAIR-2 mRNA were distinctively expressed between clinical versus subclinical hypothyroid patients and discriminated both conditions in ROC
analysis, indicating their prognostic potential. However, the limitations of this study shouldn’t be neglected. The relatively small sample size of the study investigations may limit the
interpretation of the results. Selection bias may be evident for patients as we collected the different samples from one hospital. The study results should be interpreted with caution when
extended to other populations. As a result, the findings of this study need to be validated on a bigger scale or in a community with a diverse range of racial groups. CONCLUSION This study
is the first to propose the _LncRNA-PAX8-AS1_ and _LAIR-2_ variants as novel genetic biomarkers of hypothyroidism, which could alter the LncRNA-PAX8-AS1 and LAIR-2 expression. Furthermore,
the serum LncRNA-PAX8-AS1 and LAIR-2 mRNA and protein levels have the potential as novel diagnostic and prognostic indicators of hypothyroidism. Our findings could be implemented in
hypothyroidism screening, genetic treatment, and have the possibility of large-scale application. Finally, the relationship between the studied parameters and the environmental risk factors
of hypothyroidism should be investigated. SUBJECTS AND METHODS SUBJECTS This study included 305 participants classified as 95 healthy controls and 210 hypothyroid patients recruited from the
Internal Medicine Department and Outpatient Endocrine Clinic, Kasr Al-Ainy Hospital, Cairo University from June 2020 to June 2021. Patients were classified into 100 clinical hypothyroid
cases and 110 cases with subclinical hypothyroidism depending on serum levels of TSH, free T4, and free T3 and the diagnosis was affirmed by clinical examinations. Full history taking and
clinical assessment were done for all recruited patients. Thyroid function tests were carried out, including serum levels of TSH, free T4, free T3 as well as thyroid volume. Furthermore, the
lipid profile parameters [total cholesterol, LDL-cholesterol, HDL-cholesterol and triglycerides] and the glycemic control status of patients [fasting plasma glucose, 2HPP, fasting plasma
insulin, and blood HbA1c] were assessed. Patients whose age above 18 years old from both sexes were included in the study, while those having any current malignancies, severe liver and
kidney diseases, other autoimmune diseases (except autoimmune hypothyroidism), active infection, and on any medications including glucocorticoids, hormones and drugs that interfere with
thyroid function were excluded. Clinical hypothyroid patients (92 females/8 males, age range 24–59 years) exhibited elevated serum TSH with low serum free T4 and free T3 levels. Subclinical
hypothyroid patients (104 females/6 males, age range 25–58 years) had elevated serum TSH along with normal free thyroid hormones levels. The healthy control group was age- and sex-matched to
the patient groups. The sample size was computed with a power of 0.8 as previously described by Hong and Park, 201237 using the additive model while considering the following assumptions:
case–control ratio of 1:1; 5% error rate in an allelic test; MAF; odds ratio of 2; complete linkage disequilibrium; and the prevalence of disease from published data in Egypt38. All
participants signed an informed consent. The Research and Ethics Committee for Experimental and Clinical Studies at Faculty of Pharmacy, Cairo University, Egypt approved the study protocol
and informed consent (approval number: BC29150) which conformed to the ethical guidelines of Helsinki Declaration. SAMPLE COLLECTION AND STORAGE Six milliliter blood samples were taken from
each participant and separated into two vacutainers. The first 3 mL of blood were collected into EDTA vacutainer tubes for DNA extraction and genotyping and were stored at − 80 °C until
used. The other 3 mL of the blood were stored in yellow gel vacutainer tubes, kept at room temperature for 30 min then centrifuged at 4000 rpm for 10 min to detach the sera from the clotted
whole blood. The first aliquoted sera were used for RNA extraction, whereas the other aliquoted sera were used for the LAIR-2 protein assay. Until used, all serum aliquots were maintained
frozen at a temperature of − 80 °C. DNA EXTRACTION AND GENOTYPING Genomic DNA was deduced from whole EDTA blood samples of all subjects using QIAamp DNA MiniKit as depicted in the
manufacturer's instructions (Qiagen, Valenica, CA). The yield concentration and purity were determined by NanoDrop2000 (ThermoFisher Scientific,Waltham, MA, USA). Pre-designed
primer/probe sets for the _LncRNA-PAX8-AS1_ SNPs: rs4848320 (C/T) [Assay ID: C_1940037_20, Catalog number: 4351379] and rs1110839 (G/T) [Assay ID: C_1940012_20, Catalog number: 4351379], and
the _LAIR-2_ SNP rs2287828 (C/T) [Assay ID: C_16183138_20, Catalog number: 4351379] (Applied Biosystems) were used for genotyping using the TaqMan allelic discrimination assay. DNA
amplification was done using 12.5 μL TaqMan master mix, 1.25 μL primers/probes, 1 μL DNA, and 10.25 μL H2O in a final volume of 25 μL. The real-time PCR was conducted using the Rotor gene Q
system (Qiagen) under the following cycling conditions: 95 °C for 10 min, followed by 40 cycles at 92 °C for 15 s and 60 °C for 90 s. At the end point of each cycle, fluorescence was
assessed. ASSAY OF SERUM LNCRNA-PAX8-AS1 AND LAIR 2 MRNA BY REVERSE TRANSCRIPTASE-QUANTITATIVE POLYMERASE CHAIN REACTION (RT-QPCR) Two hundred microliters of serum were used for total RNA
extraction using the miRNeasy extraction kit (Qiagen, Valenica, CA) with the supplied QIAzol lysis reagent as instructed by the manufacturer. The extracted RNA was used for both lncRNA and
mRNA expression analysis after evaluation of the concentration and purity using the NanoDrop 2000 model (ThermoFisher Scientific, USA). Reverse transcription (RT) was carried out on 0.1 μg
of total RNA in a final volume 20 μL RT reactions using RT2 First Strand kit as recommended by the manufacturer (Qiagen, Valenica, CA). Dilution of the RT product was performed with 50 μL of
RNase-free water and the diluted cDNA was stored at − 20 °C until analysis. The Maxima SYBR Green PCR kit (Thermo Fisher Scientific, Waltham, MA, USA) was used in qPCR along with GAPDH as a
housekeeping gene with ready-made primers (Qiagen Valenica, CA) for human LncRNA-PAX8-AS1 (Catalog No: 330701, specific number: LPH21217A-200), LAIR-2 mRNA (Catalog No: 249900, specific
number: QT00009499) and GAPDH (Catalog No: 249900, specific number: QT00079247). In brief, 2.5 μL appropriately diluted cDNA template was mixed with 5.5 μL RNase free water, 10 μL Maxima
SYBR Green PCR Master Mix and 2 μL ready-made forward and reverse primers in a final 20 μL reaction mixture. RT-qPCR was conducted by applying the following conditions: 95 °C for 10 min
followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C using a Rotor Gene Q system (Qiagen). A melting curve analysis was performed to ensure the specificity of PCR products of the studied
genes. 2−ΔCt was employed to compute gene expression relative to the internal control and 2−ΔΔCt was used to calculate the fold change relative to the healthy control group. ASSESSMENT OF
SERUM LAIR-2 PROTEIN A human LAIR-2 ELISA kit (E2299Hu) was obtained from Bioassay Technology Laboratory (Shanghai, China).for the quantitative measurement of serum LAIR-2 protien levels.
STATISTICAL ANALYSIS SPSS software (Chicago, IL, USA) version 25 and GraphPad Prism 8.0 statistical software (San Diego, CA, USA) were used for data analysis. Data are presented as the
median (25%-75% percentiles), mean ± standard deviation (SD) or number (percentage) when appropriate. Kolmogorov–Smirnov and Shapiro–Wilk tests were used for testing normality. Normally
distributed data were analyzed by unpaired student t test or one way ANOVA followed by Tukey’s post-hoc test when appropriate. The Mann–Whitney U test or Kruskal–Wallis test followed by
Dunn’s post-hoc test were applied to analyze the non-normally distributed data when appropriate. Chi square or Fisher’s exact test were applied to compare the categorical data. The
diagnostic and prognostic accuracy of the studied parameters were computed from the ROC curve analysis which calculates the AUC. When AUC is between 0.7 and 0.89, a potential or promising
discriminator is considered, if AUC ≥ 0.9 an excellent discriminator is considered. The correlation between the measured parameters was evaluated using Spearman's rho coefficient. To
find predictor variables linked to the risk of being diagnosed with clinical or subclinical hypothyroidism, univariate and multivariate logistic regression analyses were conducted.
Significant predictor variables from the univariate analysis were incorporated in a stepwise forward multivariate analysis (_P_ < 0.05 for inclusion and _P_ < 0.1 for exclusion from
the model). Age and sex were included as covariates to adjust the data for confounding factors. SNP analysis and the association of SNPs with clinical and subclinical hypothyroid risk were
tested using logistic regression models controlling for age and sex by employing the SNPStats online software (InistitutCatalà d’Oncologia, Barcelona, Spain;
https://www.snpstats.net/start.htm). The reference category was set as the major allele or the major homozygote genotype in the control population. The best fit model of each SNP was chosen
when it has the lowest Akaike information criterion (AIC) and the Bayesian information criterion (BIC) values compared to other genetic models. Statistical significance was set as _P_ <
0.05 for all tests, with a 95% CI. ETHICS APPROVAL AND CONSENT TO PARTICIPATE All participants signed an informed consent. The Research and Ethics Committee for Experimental and Clinical
Studies at Faculty of Pharmacy, Cairo University, Egypt approved the study protocol and informed consent (approval number: BC29150) which conformed to the ethical guidelines of Helsinki
Declaration. DATA AVAILABILITY All data generated or analyzed during this study are included in this published article and its supplementary information files. REFERENCES * Jiang, Q. _et
al._ Identification of related long non-coding RNAs and mRNAs in subclinical hypothyroidism complicated with type 2 diabetes by transcriptome analysis—A preliminary study.
_EndokrynologiaPolska_ 71, 213–226. https://doi.org/10.5603/ep.a2020.0025 (2020). Article CAS Google Scholar * Taylor, P. _et al._ Global epidemiology of hyperthyroidism and
hypothyroidism. _Nat. Rev. Endocrinol._ 14, 301–316. https://doi.org/10.1038/nrendo.2018.18 (2018). Article Google Scholar * Effraimidis, G., Watt, T. & Feldt-Rasmussen, U.
Levothyroxine therapy in elderly patients with hypothyroidism. _Front. Endocrinol._ 12, 150. https://doi.org/10.3389/fendo.2021.641560 (2021). Article Google Scholar * Peirce, C. _et al._
Treatment of refractory and severe hypothyroidism with sublingual levothyroxine in liquid formulation. _Endocrine_ 60, 193–196. https://doi.org/10.1007/s12020-017-1367-5 (2018). Article CAS
Google Scholar * Vanderpump, P. & Tunbridge, G. Epidemiology and prevention of clinical and subclinical hypothyroidism. _Thyroid_ 12, 839–847.
https://doi.org/10.1089/105072502761016458 (2002). Article Google Scholar * Eriksson, N. _et al._ Novel associations for hypothyroidism include known autoimmune risk loci. _PLoS ONE_ 7,
e34442. https://doi.org/10.1371/journal.pone.0034442 (2012). Article ADS CAS Google Scholar * Taylor, P. N. _et al._ Consortium U. Whole-genome sequence-based analysis of thyroid
function. _Nat. Commun._ 6, 1–11. https://doi.org/10.1038/ncomms6681 (2015). Article CAS Google Scholar * Shaker, O. G., Senousy, M. A. & Elbaz, E. M. Association of rs6983267 at
8q24, HULC rs7763881 polymorphisms and serum lncRNAs CCAT2 and HULC with colorectal cancer in Egyptian patients. _Sci. Rep._ 7, 1–11. https://doi.org/10.1038/s41598-017-16500-4 (2017).
Article CAS Google Scholar * Ewida, H. A., Zayed, R. K., Darwish, H. A. & Shaheen, A. A. Circulating lncRNAs HIF1A-AS2 and LINLK-A: Role and relation to hypoxia-inducible factor-1α in
cerebral stroke patients. _Mol. Neurobiol._ 58, 4564–4574. https://doi.org/10.1007/s12035-021-02440-8 (2021). Article CAS Google Scholar * Shen, Y. _et al._ Long noncoding RNA and mRNA
expression profiles in the thyroid gland of two phenotypically extreme pig breeds using ribo-zero RNA sequencing. _Genes_ 7, 34. https://doi.org/10.3390/genes7070034 (2016). Article CAS
Google Scholar * Kung, T., Colognori, D. & Lee, T. Long noncoding RNAs: Past, present, and future. _Genetics_ 193, 651–669. https://doi.org/10.1534/genetics.112.146704 (2013). Article
CAS Google Scholar * Knoll, M., Lodish, F. & Sun, L. Long non-coding RNAs as regulators of the endocrine system. _Nat. Rev. Endocrinol._ 11, 151–160.
https://doi.org/10.1038/nrendo.2014.229 (2015). Article CAS Google Scholar * Simone, R. _et al._ Serum LAIR-2 is increased in autoimmune thyroid diseases. _PLoS ONE_ 8, e63282.
https://doi.org/10.1371/journal.pone.0063282 (2013). Article ADS CAS Google Scholar * Han J, Zhou W, Jia M, Wen J, Jiang J, Shi J, Zhang K, Ma H, LiuJ, Ren J, Dai M, Hu Z, Hang D, Li
N,Shen H. Expression quantitative trait loci in long non-coding RNA PAX8-AS1 are associated with decreased risk of cervical cancer. Mol. Genet. Genom. 2016;291:1743–1748.
https://doi.org/10.1007/s00438-016-1217-9. * Macchia, E. _et al._ PAX8 mutations associated with congenital hypothyroidism caused by thyroid dysgenesis. _Nat. Genet._ 19, 83–86.
https://doi.org/10.1038/ng0598-83 (1998). Article CAS Google Scholar * Fernandez, P., Marquez, A. & Santisteban, P. Thyroid transcription factors in development, differentiation and
disease. _Nat. Rev. Endocrinol._ 11, 29–42. https://doi.org/10.1038/nrendo.2014.186 (2015). Article CAS Google Scholar * Nica, C. & Dermitzakis, T. Expression quantitative trait loci:
Present and future. _Philos. Trans. R. Soc. B Biol. Sci._ 368, 20120362. https://doi.org/10.1098/rstb.2012.0362 (2013). Article CAS Google Scholar * Veyrieras, B. _et al._
High-resolution mapping of expression-QTLs yields insight into human gene regulation. _PLoS Genet._ 4, e1000214. https://doi.org/10.1371/journal.pgen.1000214 (2008). Article CAS Google
Scholar * Bahari, G., Hashemi, M., Naderi, M., Sadeghi-Bojd, S. & Taheri, M. Long non-coding RNA PAX8-AS1 polymorphisms increase the risk of childhood acute lymphoblastic leukemia.
_Biomed. Rep._ 8, 184–190. https://doi.org/10.3892/br.2017.1028 (2018). Article CAS Google Scholar * Lebbink RJ, van den Berg MC, de Ruiter T, Raynal N, van Roon JA, Lenting PJ, Jin
B,Meyaard L. The soluble leukocyte-associated Ig-like receptor (LAIR)-2 antagonizes the collagen/LAIR-1 inhibitory immune interaction. _J. Immunol._ 2008;180:1662–1669.
https://doi.org/10.4049/jimmunol.180.3.1662. * Lebbink, J. _et al._ Identification of multiple potent binding sites for human leukocyte associated Ig-like receptor LAIR on collagens II and
III. _Matrix Biol._ 28, 202–210. https://doi.org/10.1016/j.matbio.2009.03.005 (2009). Article CAS Google Scholar * Camargo, M., Augusto, G. & Petzl-Erler, L. Differential gene
expression levels might explain association of LAIR2 polymorphisms with pemphigus. _Hum. Genet._ 135, 233–244. https://doi.org/10.1007/s00439-015-1626-6 (2016). Article CAS Google Scholar
* Díaz-Peña, R. _et al._ A high density SNP genotyping approach within the 19q13 chromosome region identifies an association of a CNOT3 polymorphism with ankylosing spondylitis. _Ann.
Rheum. Dis._ 71, 714–717. https://doi.org/10.1136/annrheumdis-2011-200661 (2012). Article CAS Google Scholar * Jabbar, A. _et al._ Thyroid hormones and cardiovascular disease. _Nat. Rev.
Cardiol._ 14, 39–55. https://doi.org/10.1038/nrcardio.2016.174 (2017). Article CAS Google Scholar * Apostu, D. _et al._ The influence of thyroid pathology on osteoporosis and fracture
risk: A review. _Diagnostics_ 10, 149–160. https://doi.org/10.3390/diagnostics10030149 (2020). Article CAS Google Scholar * Ichiki, T. Thyroid hormone and atherosclerosis. _Vascul.
Pharmacol._ 52, 151–156. https://doi.org/10.1016/j.vph.2009.09.004 (2010). Article CAS Google Scholar * Matana, A. _et al._ Genome-wide meta-analysis identifies novel gender specific loci
associated with thyroid antibodies level in Croatians. _Genomics_ 111, 737–743. https://doi.org/10.1016/j.ygeno.2018.04.012 (2019). Article CAS Google Scholar * Castanet, M. _et al._
Thyroid hemiagenesis is a rare variant of thyroid dysgenesis with a familial component but without Pax8 mutations in a cohort of 22 cases. _Pediatr. Res._ 57, 908–913.
https://doi.org/10.1203/01.pdr.0000161409.04177.36 (2005). Article CAS Google Scholar * Zhou, P., Xu, T., Hu, H. & Hua, F. Overexpression of PAX8-AS1 inhibits malignant phenotypes of
papillary thyroid carcinoma cells via miR-96-5p/PKN2 axis. _Int. J. Endocrinol._ 2021, 1–16. https://doi.org/10.1155/2021/5499963 (2021). Article CAS Google Scholar * Roy, P., Orecchioni,
M. & Ley, K. How the immune system shapes atherosclerosis: roles of innate and adaptive immunity. _Nat. Rev. Immunol._ 22, 251–265. https://doi.org/10.1038/s41577-021-00584-1 (2021).
Article CAS Google Scholar * Abdelazim, S. A., Shaker, O. G., Aly, Y. A. H. & Senousy, M. A. Uncovering serum placental-related non-coding RNAs as possible biomarkers of preeclampsia
risk, onset and severity revealed MALAT-1, miR-363 and miR-17. _Sci. Rep._ 12, 1249. https://doi.org/10.1038/s41598-022-05119-9 (2022). Article ADS CAS Google Scholar * Motawi, T. K.,
Shaker, O. G., Hassanin, S. O., Ibrahim, S. G. & Senousy, M. A. Genetic and epigenetic control on clock genes in multiple sclerosis. _J. Genet. Genom._ 49, 74–76.
https://doi.org/10.1016/j.jgg.2021.07.016 (2022). Article Google Scholar * Radwan, A. F., Shaker, O. G., El-Boghdady, N. A. & Senousy, M. A. Association of MALAT1 and PVT1 variants,
expression profiles and target miRNA-101 and miRNA-186 with colorectal cancer: Correlation with epithelial–mesenchymal transition. _Int. J. Mol. Sci._ 22, 6147.
https://doi.org/10.3390/ijms22116147 (2021). Article CAS Google Scholar * Senousy, M. A., El-Abd, A. M., Abdel-Malek, R. R. & Rizk, S. M. Circulating long non-coding RNAs HOTAIR,
Linc-p21, GAS5 and XIST expression profiles in diffuse large B-cell lymphoma: Association with R-CHOP responsiveness. _Sci. Rep._ 11, 2095. https://doi.org/10.1038/s41598-021-81715-5 (2021).
Article CAS Google Scholar * Senousy, M. A., Shaker, O. G., Sayed, N. H., Fathy, N. & Kortam, M. A. LncRNA GAS5 and miR-137 polymorphisms and expression are associated with multiple
sclerosis risk: Mechanistic insights and potential clinical impact. _ACS ChemNeurosci._ 11, 1651–1660. https://doi.org/10.1021/acschemneuro.0c00150 (2020). Article CAS Google Scholar *
Abd-Elmawla, M. A., Hassan, M., Elsabagh, Y. A., Alnaggar, A. R. L. R. & Senousy, M. A. Deregulation of long noncoding RNAs ANCR, TINCR, HOTTIP and SPRY4-IT1 in plasma of systemic
sclerosis patients: SPRY4-IT1 as a novel biomarker of scleroderma and its subtypes. _Cytokine_ 133, 155124. https://doi.org/10.1016/j.cyto.2020.155124 (2020). Article CAS Google Scholar *
Hong, E. P. & Park, J. W. Sample size and statistical power calculation in genetic association studies. _Genomics Inform._ 10(2), 117–122. https://doi.org/10.5808/GI.2012.10.2.117
(2012). Article MathSciNet Google Scholar * El-Hadidi, K. T., Mansour, M. A. & El-Wakd, M. M. Thyroid dysfunction and anti-thyroid antibodies in Egyptian patients with systemic lupus
erythematosus: Correlation with clinical musculoskeletal manifestations. _Egypt. Rheumatol._ 364, 173–178. https://doi.org/10.1016/j.ejr.2014.01.006 (2014). Article Google Scholar Download
references ACKNOWLEDGEMENTS The authors are grateful for all participants in the work and technicians who helped in carrying out the practical experiments. FUNDING Open access funding
provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This paper is based upon work supported by Science,
Technology & Innovation Funding Authority (STDF) under Grant Number 44393. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Egyptian Drug Authority, Cairo, Egypt Omar M. Elsayed *
Biochemistry Department, Faculty of Pharmacy, Cairo University, Cairo, 11562, Egypt Samy A. Abdelazim & Mahmoud A. Senousy * Pharmacology, Toxicology and Biochemistry Department, Faculty
of Pharmacy, Future University in Egypt (FUE), Cairo, Egypt Hebatallah A. Darwish * Medical Biochemistry and Molecular Biology Department, Faculty of Medicine, Cairo University, Cairo,
Egypt Olfat G. Shaker * Department of Biochemistry, Faculty of Pharmacy and Drug Technology, Egyptian Chinese University, Cairo, 11786, Egypt Mahmoud A. Senousy Authors * Omar M. Elsayed
View author publications You can also search for this author inPubMed Google Scholar * Samy A. Abdelazim View author publications You can also search for this author inPubMed Google Scholar
* Hebatallah A. Darwish View author publications You can also search for this author inPubMed Google Scholar * Olfat G. Shaker View author publications You can also search for this author
inPubMed Google Scholar * Mahmoud A. Senousy View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS O.M.E participated in the investigation,
resources, methodology, statistical analysis, funding acquisition, visualization, and writing of the original draft. S.A.A and H.A.D conceptualized and supervised the work and participated
in writing, review & editing. O.G.S participated in the investigation, sample collection, and methodology. M.A.S participated in the investigation, statistical analysis, data curation,
data interpretation, validation, visualization, and writing of the original draft. All authors have reviewed and approved the final manuscript. CORRESPONDING AUTHOR Correspondence to Samy A.
Abdelazim. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under
a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate
credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article
are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons
licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of
this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Elsayed, O.M., Abdelazim, S.A., Darwish, H.A. _et al._
Association of LncRNA-PAX8-AS1 and LAIR-2 polymorphisms along with their expression with clinical and subclinical hypothyroidism. _Sci Rep_ 13, 6 (2023).
https://doi.org/10.1038/s41598-022-26346-0 Download citation * Received: 29 August 2022 * Accepted: 13 December 2022 * Published: 02 January 2023 * DOI:
https://doi.org/10.1038/s41598-022-26346-0 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative