Crispr/cas9 editing of the myo7a gene in rhesus macaque embryos to generate a primate model of usher syndrome type 1b
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Mutations in the _MYO7A_ gene lead to Usher syndrome type 1B (USH1B), a disease characterized by congenital deafness, vision loss, and balance impairment. To create a nonhuman
primate (NHP) USH1B model, CRISPR/Cas9 was used to disrupt _MYO7A_ in rhesus macaque zygotes. The targeting efficiency of Cas9 mRNA and hybridized crRNA-tracrRNA (hyb-gRNA) was compared to
Cas9 nuclease (Nuc) protein and synthetic single guide (sg)RNAs. Nuc/sgRNA injection led to higher editing efficiencies relative to mRNA/hyb-gRNAs. Mutations were assessed by preimplantation
genetic testing (PGT) and those with the desired mutations were transferred into surrogates. A pregnancy was established from an embryo where 92.1% of the PGT sequencing reads possessed a
single G insertion that leads to a premature stop codon. Analysis of single peripheral blood leukocytes from the infant revealed that half the cells possessed the homozygous single base
insertion and the remaining cells had the wild-type _MYO7A_ sequence. The infant showed sensitive auditory thresholds beginning at 3 months. Although further optimization is needed, our
studies demonstrate that it is feasible to use CRISPR technologies for creating NHP models of human diseases. SIMILAR CONTENT BEING VIEWED BY OTHERS PREVENTING AUTOSOMAL-DOMINANT HEARING
LOSS IN _BTH_ MICE WITH CRISPR/CASRX-BASED RNA EDITING Article Open access 14 March 2022 PRODUCTION OF A HETEROZYGOUS EXON SKIPPING MODEL OF COMMON MARMOSETS USING GENE-EDITING TECHNOLOGY
Article Open access 30 August 2024 AAV-MEDIATED _FOXG1_ GENE EDITING IN HUMAN RETT PRIMARY CELLS Article 15 June 2020 INTRODUCTION USH is the leading cause of congenital deaf-blindness in
humans with a prevalence of 4 to 17 per 100,0001,2. This syndromic form of retinitis pigmentosa (RP) is characterized by deficits in hearing, balance and vision3,4,5. Depending on the
disease phenotype and severity of symptoms, USH can be classified into three subtypes. USH1 patients show congenital deafness, vestibular dysfunction, and the onset of retinal degeneration
within 10 years from birth. USH2 patients show moderate to severe congenital deafness and their onset of RP is delayed until late adolescence. USH3 patients show gradual hearing loss and
variable onset of RP4,5. Mutations in the _MYO7A_ gene are a cause of USH1B. The protein encoded by the _MYO7A_ gene, myosin VIIA, is an unconventional myosin motor protein expressed in
several tissues, including the inner ear and eye5. Gene augmentation therapy and cell transplantation are potential treatments for individuals with USH, including USH1B. Gene augmentation
therapy is challenging due to the large size of the _MYO7A_ gene, requiring the development of dual-AAV vectors and other strategies for gene delivery6,7,8. While initial results with
dual-AAV vectors are promising, questions still persist about the design of optimal gene therapies for replacement of large genes such as _MYO7A_, timing of treatment, proper
immunosuppression, and the optimal surgical method for delivery of gene therapy vectors or transplanted cells in humans9,10. Within the inner ear, several gene therapy strategies were
successfully utilized in mouse models with cochlear hair cell defects; however, it is predicted that these interventions would need to be delivered prenatally in humans11. Accordingly, an
animal model with greater translatability to humans is necessary to further pursue the full range of potential therapeutic technologies. Rhesus macaques share a high degree of genetic,
physiological, anatomical, and behavioral similarities with humans. As such, they represent an ideal human surrogate for testing and optimizing new therapies. NHP models of Usher syndrome
are particularly urgently needed because rodent models of USH1 fail to show a retinal degeneration phenotype due to differences in the cellular distribution of _MYO7A_ expression. In humans
and NHPs, _MYO7A_ is expressed in pigment epithelium cells and photoreceptors. _MYO7A_ expression in the mouse only occurs in pigment epithelium, which likely explains the lack of
significant vision loss in _Myo7a_ null mutant mice12,13. However, the production of transgenic NHPs to serve as human disease models has largely been unfeasible due to inefficiencies in
technologies such as somatic cell nuclear transfer or chimeric NHP production using stem cells14,15,16,17. Through the use of the programmable nuclease CRISPR/Cas9, these issues can be
circumvented by direct injection of Cas9 and the relevant gRNA into zygotes. While injection of CRISPR/Cas9 in NHP embryos has shown promise in creating disease models, using such an
approach still faces significant hurdles18. These include optimizing injection reagents and injection timing for maximum gene targeting efficiency while prescreening transferable embryos for
the desired edit. A previous study demonstrated that simultaneous delivery of three gRNAs targeting regions in close proximity to one another, as compared to a single gRNA, increased the
creation of insertion or deletion (indel) mutations leading to a higher knockout (KO) efficiency in cynomolgus macaque embryos19. Moreover, different formulations of gRNAs and Cas9 (i.e.,
mRNA versus protein) are available that may impact embryo editing efficiencies. A commonly used gRNA system includes a Hyb-gRNA formulation that requires annealing of crRNA with a tracrRNA
to form a functional gRNA. In contrast, synthetic single gRNAs possess both Cas9 binding and target gene protospacer sequences, typically incorporating endonuclease resistant nucleotides
that prevent its degradation. Currently, limited information exists regarding the efficacy of these different formulations of gRNA/Cas9 for editing rhesus macaque embryos. To generate an
USH1B NHP model, exon 3 of the rhesus macaque _MYO7A_ gene was targeted to introduce indels. Exon 3 codes for the region of the protein that is upstream of the motor domain of myosin VIIA,
which is a major site of pathogenic mutations in humans20,21. After confirmation of gRNA efficiency in vitro in the rhesus macaque CMMT cell line, CRISPR/Cas9 reagents were injected into
rhesus macaque zygotes. Our results demonstrate that Nuc/sgRNA injection possessed significantly greater _MYO7A_ editing rates as determined in trophectoderm (TE) biopsies and arrested
embryos relative to the injection of mRNA/hyb-gRNA. However, the use of Nuc/sgRNA resulted in a significantly lower blastocyst development rate relative to the mRNA/hyb-gRNA injections. Six
embryos were transferred to five surrogates, resulting in the birth of one _MYO7A_ KO female macaque named “Mya”. Single-cell genotyping results showed that Mya carries a mosaic mutation
within exon 3 of the _MYO7A_ gene. Auditory thresholds at 3–12 months were consistent with age-matched controls, and retinal structure and function also appeared normal at all ages tested.
These results demonstrate the feasibility of a gene-editing approach and assisted reproductive technologies for the creation of valuable NHP disease models, although further studies are
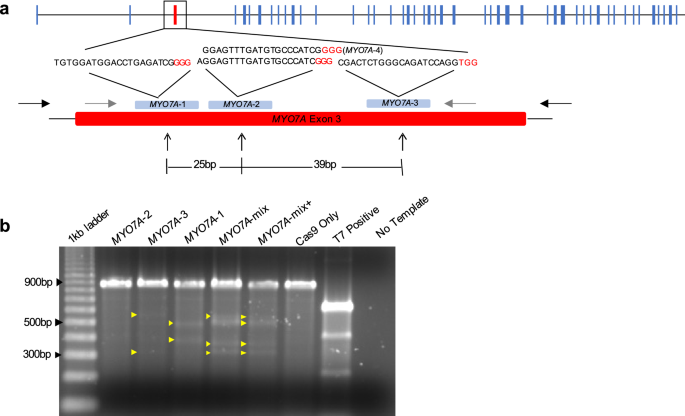
needed to improve overall efficiencies, reduce the incidence of mosaicism, and produce live births of fully edited animals. RESULTS IN VITRO VALIDATION OF _MYO7A_ GRNAS Four gRNAs targeting
exon 3 of _MYO7A_ were developed and initially tested for their targeting efficiency in vitro. Cas9 protein and each hyb-gRNA were combined with PCR amplicons containing the wild-type
_MYO7A_ exon 3 target site. All four gRNAs successfully generated double-stranded DNA breaks (DSB) on wild-type _MYO7A_ amplicons (Supplementary Fig. 1). Three out of four hyb-gRNAs (#1
through #3) were subsequently tested in rhesus macaque mammary gland CMMT cells and #4 was excluded because the target sequences of gRNAs #2 and #4 overlapped by 19 bp (Fig. 1a). In CMMT
cells, hyb-gRNA #1 and #3 resulted in the generation of indels in the _MYO7A_ gene based on a T7E1 assay. Limited editing was observed with hyb-gRNA #2. However, when hyb-gRNA #1, #2, and #3
were co-transfected with a Cas9 expressing plasmid into CMMT cells, robust indel formation in _MYO7A_ exon 3 was observed (Fig. 1b). ASSESSING TARGETING EFFICIENCY OF DIFFERENT CAS9 AND
GRNA PLATFORMS IN RHESUS MACAQUE EMBRYOS Based on the validation of editing in CMMT cells, hyb-gRNAs comprised of a mixture of gRNAs 1 through 3 and Cas9 mRNA were injected into rhesus
macaque zygotes (N = 53) to introduce indels in the _MYO7A_ gene (Table 1). Out of 11 (20.75%) blastocysts and 33 (62.26%) arrested embryos, 36 samples (8 biopsies, 28 arrested embryos)
successfully underwent whole genome amplification (WGA) and were genotyped by next-generation sequencing (NGS). NGS analysis showed 38.8% of embryos contained at least one edited allele.
Nuc/sgRNAs for target sites #1, #2, and #3 were subsequently injected in a total of 234 embryos, of which 200 (85.47%) were arrested prior to forming a blastocyst and 26 (11.11%) became
blastocysts. A total of 211 samples (19 biopsies, 192 arrested cleavage stage embryos) underwent successful WGA and were genotyped. Blastocyst biopsies and arrested embryos obtained after
Nuc/sgRNA protein injection had an indel rate that was higher than that of mRNA/hyb-gRNA injected embryos (76.3% versus 38.8%, respectively; p < 0.01). Of the embryos observed to possess
_MYO7A_ indels after Nuc/sgRNA injection, a higher percentage (65.21%) were fully edited (i.e., no wild-type sequence detected) relative to edited embryos obtained following hyb-gRNA/Cas9
mRNA injection (28.5%; p < 0.01) (Table 1). Injection of Nuc/sgRNA resulted in a higher embryo arrest rate than mRNA/hyb-gRNAs injection (p < 0.0011; Table 1). Although the injection
of mRNA/hyb-gRNA resulted in a blastocyst formation rate (20.75%) that was higher than that observed using Nuc/sgRNA (11.11%), the difference was not statistically significant (p = 0.0697).
TRANSFER OF EDITED RHESUS MACAQUE EMBRYOS All blastocysts were cryopreserved following trophectoderm biopsy. Following biopsy genotyping, one blastocyst obtained from mRNA/hyb-gRNA injection
(embryo 190; E190) and one from Nuc/sgRNA injection (E773) were transferred into one recipient female rhesus macaque. DNA sequencing of the trophectoderm biopsy revealed that 92.1% of the
sequencing reads for E190 possessed a single G insertion that would lead to a frameshift mutation. Sequencing of DNA obtained from the trophectoderm biopsy of E773 revealed that 98.5% of the
sequencing reads possessed a 17 bp insertion/50 bp deletion, which would lead to an in-frame 18 bp deletion (Fig. 2a). Additional single embryo transfers (embryos E180, E1170, E1171, and
E1289) were performed (n = 4). TE cell analysis showed that E180 carried 99.6% of two different mutations resulting in premature stop codons and 0.2% of wild-type sequence. Embryos, E1170,
E1171, and E1289, did not carry any wild-type sequence and all contained frameshift mutations (Supplementary Fig. 2). Out of 5 embryo transfers, a successful singleton pregnancy resulted
from the transfer of E190 and E773 into one recipient (20% live birth rate per transfer; Fig. 2b and Supplementary Table 1). A female infant (“Mya”) was delivered by C-section at 150 days
gestation (typical gestational length is 158 to 173 days). Mya’s birth weight was 390 g, which was lower than the full-term average of 488 g22. Within minutes of delivery, Mya was breathing
on her own and appeared healthy (Fig. 3a). Four different tissue samples, including placenta, blood, skin, and buccal specimens, were collected at or within 3 weeks after birth to identify
the origin of the transferred embryo based on the _MYO7A_ mutation and to assess the level of editing in the offspring relative to what was observed in the TE biopsy (Fig. 3b). The
genotyping results from collected samples showed a ‘G’ insertion, confirming that the pregnancy was the result of implantation of E190. In the tissues sampled, the percentage of mutant
sequence relative to wild-type _MYO7A_ sequence was 40–50%, depending on the tissue analyzed (two biological replicates), compared to the 92.1% mutant _MYO7A_ sequence observed in the TE
biopsy. To ensure that a larger deletion of the entire exon 3 did not occur, which would be missed by the PCR primers that recognize the sequence directly adjacent to the gRNA target region,
primers were used to amplify the entire _MYO7A_ exon 3 and adjacent introns in genomic DNA obtained from Mya. No larger deletions encompassing all of exon 3 were observed (Supplementary
Fig. 3). Moreover, only a single edit (1 bp insertion) was observed from the sequencing results of the larger exon 3 amplicon, further suggesting additional larger deletions (i.e., deletion
of exon 3) did not occur. The observed level of editing being close to 50% could be due to either editing of one _MYO7A_ allele in the one-cell stage (e.g., heterozygous) or homozygous
editing in one cell of a 2-cell embryo (e.g., homozygous mosaic). To distinguish between these possibilities, single leukocytes were isolated from a blood sample by FACS and analyzed for the
presence or absence of the introduced G insertion following WGA, PCR, and Sanger sequencing. A total of 13 individual cells (59%) were homozygous for the G insertion, whereas 9 cells (41%)
had wild-type sequence, confirming that Mya is mosaic for a homozygous frameshift mutation in the _MYO7A_ gene (Fig. 3c). ASSESSMENT OF OFF-TARGET EDITING To determine if off-target editing
occurred, regions of the rhesus macaque genome with the highest level of homology to the _MYO7A_ gRNA recognition sites were chosen for analysis, where each potential editing site had 2–4 bp
mismatches relative to the target sequence of each of the gRNAs (Supplementary Table 2). Based on these criteria, 9 sites in the rhesus macaque genome were chosen for further analysis. DNA
was extracted from blood, skin, placenta, and buccal cells collected from Mya. Putative off-target sites were PCR amplified and sequenced. Sequencing results revealed that 99.3% of sequence
reads corresponded to the wild-type sequence, while < 0.7% possessed a sequence that differed from the reference _MYO7A_ sequence. However, these differences were located outside of the
sgRNA target sequence, indicating that they represent naturally occurring variants or PCR/sequencing errors. Comparing the region of the amplicon sequences that correspond to the region of
homology with the gRNAs, no detectable off-target mutations were observed at the nine sites analyzed (Supplementary Fig. 4). COMPARISON OF TARGET GENE EDITING WITHIN THE RHESUS MACAQUE
EMBRYO RELATIVE TO ITS TE BIOPSY Based on the discrepancy in the level of editing observed in the TE biopsy versus tissues obtained from Mya (92.1% versus 59%, respectively), additional
sequencing was conducted to assess the accuracy of TE biopsy sequence data in predicting the genotype of the corresponding embryo. Genotyping results between biopsied samples and blastocysts
were found to vary (Table 2). The TE biopsy of E1172 possessed 54.42% wild-type sequence reads and 41.65% sequencing reads that included a 70 bp deletion. Sequence analysis of the
corresponding blastocyst DNA revealed a lower percentage of wild-type _MYO7A_ sequence reads (21.1%). Moreover, the remaining 79.9% sequence corresponded to 8 different indels, but not the
70 bp deletion observed in the biopsied TE sample. The TE biopsy from E1851 sequencing possessed 90.65% of a 1 bp indel and 2.18% of the wild-type sequence. However, the corresponding
blastocyst sequencing possessed 42.86% wild-type sequence and 57.15% contained 3 different mutations from TE biopsy result. The sequencing results of TE cells from E1850 also possessed a
mixture of wild-type and _MYO7A_ edited sequences, of which 12.04% were wild-type sequence and 84.17% included a 1 bp insertion. The blastocyst, however, carried no wild-type sequence and
two mutations that differed from what was observed in the TE biopsy. In other instances, greater predictability of the overall level of indels in the biopsy related to the rest of the
embryo. For example, E1166 TE biopsy possessed 94.5% wild-type sequence and a mutation was not detected, whereas the remainder of the embryo had 84.84% wild-type sequence along with 3.74%
edited sequence that included a 2 bp insertion. The TE biopsy for E1285 had 52.4% wild-type sequence and 45.78% sequence reads with a 1 bp insertion, whereas the remainder of the embryo
carried 61.67% wild-type and 30.59% with 4 different mutations. Only 10.71% of the same 1 bp insertion was detected in the embryo. In E1850, the biopsied TE cells possessed 12.04% wild-type
and 84.17% of an edited _MYO7A_ sequence, whereas the blastocyst had 93.1% sequence showing two different mutations. EVALUATION OF AUDITORY FUNCTION To assess hearing function in Mya, we
measured auditory brainstem responses (ABR) and distortion product otoacoustic emissions (DPOAE) measures at one month of age. No ABR thresholds were detected from 0.5 to 16 kHz (Fig. 4a),
and no DPOAE responses were observed from 2 to 12 kHz (Fig. 5a), which is consistent with the phenotype predicted for a null _MYO7A_ mutation. However, 26 kHz ABR thresholds were present
bilaterally at one month, though dramatically elevated compared to controls (Fig. 4a). Similarly, DPOAE responses that increased in amplitude with increasing stimulus intensity were present
at 16 and 26 kHz (Fig. 5a). These high frequency ABR and DPOAE responses suggested that Mya retained some restricted auditory function. Subsequent longitudinal analyses indicated that Mya
had sensitive ABR thresholds from 0.5 to 26 kHz at 3, 6, and 12 months (Fig. 4) and robust DPOAE responses from 2 to 26 kHz at 3 and 12 months (Fig. 5b,c). The data indicate that Mya has
sufficient functional inner and outer hair cells that support sensitive hearing compared to age-matched controls. EVALUATION OF RETINAL STRUCTURE AND FUNCTION Multimodal retinal imaging,
conducted at 2, 4, 6, 9, and 12 months of age, included color fundus photography, spectral domain ocular coherence tomography (sdOCT), fundus autofluorescence (FAF) imaging, ultra-widefield
imaging and fluorescein and indocyanine angiography. No pathological features were observed in any imaging mode at any age. A color fundus photograph and a macular sdOCT scan for Mya at 6
months are shown in Fig. 6a,b. The thicknesses of 11 retinal layers were determined by the segmentation of sdOCT images. The only abnormality detected was in the thickness of the inner and
outer segment layer, which was slightly below the normal range throughout the central retina (Fig. 6c). Retinal function was assessed by full-field and multifocal electroretinograms (ERGs)
at 2.5, 6, 9 and 12 months. Response amplitudes and timing were similar to those of normal age-matched rhesus macaque infants in both scotopic (dark-adapted) or photopic (light-adapted)
conditions, indicating normal rod and cone photoreceptor function at each age (Supplementary Fig. 5). DISCUSSION Direct injection of Cas9 and gRNAs into rhesus macaque zygotes allows for the
creation of genetically modified NHPs in a rapid and relatively efficient manner23. Rhesus macaques have a relatively long gestational period (~ 165 days) and take 3–4 years to reach sexual
maturity. By avoiding the need for chimera generation, direct injection into zygotes would be a more efficient route for generating specific disease models. We showed that high efficiency
editing can be achieved through injection of multiple sgRNAs and Cas9 protein into the rhesus macaque zygote. By directly injecting CRISPR/Cas9 into rhesus macaque zygotes, modification of
the _MYO7A_ gene was achieved and resulted in a live-born female infant (Mya). These studies demonstrate that it is possible to target _MYO7A_ for the development of a NHP USH1B model.
However, further investigation should be conducted to maximize target gene modification efficiency. Similar targeting efficiency was observed when several _MYO7A_ specific gRNAs were tested
in a cell-free DNA cleavage assay. However, validation in rhesus macaque CMMT cells demonstrated that three different sgRNAs possessed varying editing efficiency by themselves, but appeared
more robust in generating indels in the _MYO7A_ gene when used together. These differing results from the PCR amplicon test and the cell line validation might be explained by differing
chromosomal accessibility and organization. PCR amplicons of target regions likely offer unobstructed digestion by CRISPR/Cas924. After observing efficient editing in CMMT cells with the
combination of three sgRNAs, studies were initiated to determine editing rates within rhesus macaque zygotes using different gRNA and Cas9 formulations, namely the combination of Cas9 mRNA
and a hybrid gRNA system (mRNA/hyb-gRNA) compared to Cas9 nuclease protein and a synthetic single gRNA system (Nuc/sgRNA). On-target editing efficiency in rhesus macaque zygotes with
Nuc/sgRNA injection was similar to what was reported in various species25,26,27. Alongside higher editing efficiency, we saw a greater proportion of arrested embryos in Nuc/sgRNA injected
embryos relative to those receiving mRNA/hyb-gRNA. Similar effects were observed in channel catfish, where an increased level of editing and lower embryonic developmental rates were
associated with increasing Cas9 nuclease and gRNAs concentrations28. The tradeoff in increased editing efficiency with reduced blastocyst formation rates may be due to several factors. It is
possible that more efficient on-target editing is coupled with more efficient off-target editing of embryonic lethal genes that have a negative impact on embryo development. Moreover,
recent research in human embryos revealed a propensity of CRISPR/Cas9 to cause chromosomal rearrangements and higher aneuploidy rates, which in turn may compromise embryonic development29.
It is possible that the embryo that gave rise to Mya also possessed chromosomal abnormalities. However, Mya’s fetal and postnatal development was similar to unedited control rhesus macaques,
indicating there are no substantial chromosomal rearrangements present that would be incompatible with implantation, pregnancy, or normal development. Analysis of cell biopsies from
developing embryos is commonly used to detect aneuploidy and disease-causing mutations in human IVF clinics30. To avoid wasting resources by transferring and establishing pregnancies with an
unedited or incompletely edited embryo, TE biopsies were used to determine the level of gene editing before CRISPR/Cas9 injected embryos were transferred into surrogates. However, the
genotyping of different tissues and cells obtained from Mya indicates that a TE biopsy may not fully reflect the edit or range of edits present in the rest of the embryo. The genotyping
result from the biopsied TE cells of Mya (E197) showed that 92% of the reads contained a G insertion, which would lead to a frameshift and a premature stop codon, and 8% of the sequencing
reads corresponded to wild-type _MYO7A_ sequence. Genotyping results from tissue biopsies collected from Mya, however, showed that there were different levels of mutations within different
tissues (i.e., 40% of sequence reads with the G insertion in blood cells versus approximately 50% in cells obtained from a buccal swab). These findings suggested that Mya was mosaic for
wild-type and homozygous mutant editing rather than heterozygous because the latter would be expected to give a 50% edited and 50% wild-type sequence in all tissues analyzed. Single
peripheral blood leukocytes possessed only the G insertion or wild-type _MYO7A_ sequence, thereby confirming that Mya is mosaic for the homozygous mutation. To determine if a large deletion
exists that encompasses all of _MYO7A_ exon 3, primers (888 bp) were used that amplify the entire exon 3 and portions of the flanking introns. PCR product size and Sanger sequencing
indicated there was no large deletion. The possibility of larger chromosomal insertions, deletions, or rearrangements cannot be ruled out. However, due to the normal fetal and neonatal
development and behavior of Mya, the likelihood of significant chromosomal abnormalities is low. Based on these results, it is likely that the editing event occurred at the 2-cell stage
leading to a mixture of wild-type and homozygous mutant (1 bp insertion) cells in Mya. Our observations are similar to a previously published report wherein sheep blastocysts that arose from
CRISPR/Cas9 injected zygotes were biopsied and the TE biopsy was compared to tissue samples collected from the offspring from successful embryo transfers31. Similar to our results, the
genotypes of the biopsy and the remaining embryo were less consistent when biopsies contained some intermediate level of editing. However, several embryos (E1166, E1285, and E1850) exhibited
a similar ratio of wild-type and mutation alleles in the comparison of the biopsy and remaining embryo. Based on these results, PGT may not be suitable for precise genotyping but rather
serves as a general indicator of whether any wild-type allele is present in the embryo. It is understandable that the results obtained from a TE biopsy may not fully represent the editing
that occurs in the offspring, considering that a typical TE biopsy contains 10–15 cells, which is less than 5% of the population of a whole embryo32. Another viewpoint is that if genetic
modification happens in the later stage of the embryo (2, 4, 8 cell stages), the accuracy of TE biopsy genotyping is decreased. On the other hand, if editing occurs at the zygote stage, then
TE biopsy genotyping provides a more accurate measure of editing. A limitation in our comparison test was that only 5 TE biopsy-embryo pairs were used. Thus, the analyses of additional TE
biopsies and embryos will be needed to define the accuracy of using TE biopsies for predicting the level of editing in the corresponding embryo. Off-target mutations are a major concern with
all programmable endonucleases33,34. CRISPR/Cas9 targeting of a specific gene relies on a 20 nt spacer sequence at the 5′ end of the guide RNA that is complementary to the gene of interest,
and if the genome is large and variable enough, homologous off-target regions of the genome could be affected35. Off-target editing is more likely to happen at sites with 1–5 bp mismatches
relative to the gRNA target sequence and distal to the PAM sequence36,37. However, this is not absolute because a recent study demonstrated that off-targeting events occurred when mismatches
were located at the PAM proximal end (i.e., near or adjacent to the seed sequence)38. Genomic DNA from Mya was used to assess a total of nine regions of the rhesus macaque genome that
possessed high homology to the three gRNAs that were used in the zygote injections (i.e., the 3 regions of greatest homology for each gRNA). Despite each potential off-target site only
possessing 2–3 bp mismatches, no off-targeting events were detected at these sites. It should be noted that whole genome sequencing or other large scale CRISPR off-target analyses
(CIRCLE-Seq, GUIDE-seq) were not conducted, therefore, potential off-target mutations cannot be completely ruled out. If a gene-edited model of USH1B faithfully reproduces the human disease,
we would expect profound hearing loss at birth as the first indication of an Usher disease phenotype. Therefore, quantitative assessment of auditory function, including ABR and DPOAEs, are
essential to validate hearing loss in a rhesus model. Inner hair cells of the organ of Corti forward almost all of the acoustic information collected by the cochlea to the brain39. ABRs are
a sensitive measure of auditory function that reflects inner hair cell activity40. Outer hair cells amplify low intensity sounds and the robustness of DPOAEs reflect outer hair cell
dynamics40,41,42. A null mutation in _MYO7A_ would be expected to cause gross perturbation of inner and outer hair cell structure, leading to hair cell death and the absence of ABR and DPOAE
responses. At one month, we were unable to detect ABR responses from 0.5 to 16 kHz, or DPOAE responses from 2 to 12 kHz in Mya (Fig. 5a). Surprisingly, we detected bilateral, though grossly
elevated, ABR thresholds at 26 kHz and sensitive DPOAEs at 16 and 26 kHz in Mya at one month. One explanation for the mixed hearing loss is mosaic _MYO7A_ expression in inner and outer hair
cells that differentially affects the apex through the middle turn of the cochlea, which subserves low and intermediate frequency responses43. While plausible, subsequent detection of
sensitive ABR thresholds from 0.5 to 26 kHz at 3, 6, and 12 months, and robust DPOAE responses from 2 to 26 kHz at 3 and 12 months, demonstrated that Mya had sensitive hearing compared to
age-matched controls. If Mya is mosaic for _MYO7A_ expression in sensory hair cells, the extent of hair cell compromise is insufficient to grossly affect auditory function. Although
unedited, full-term controls exhibited normal auditory responses at one month of age, the inability to detect ABR and DPOAE responses in Mya may be anatomic or developmental. Since Mya was
born 15 days early, the lack of response may be more related to the narrowness of the neonatal rhesus macaque ear canal that complicates acoustic probe placement. In this constraining
anatomical context, it cannot be ruled out that inefficient transmission of pure tone stimuli may have confounded both the ABR and DPOAE measures at one month of age. Most forms of retinitis
pigmentosa, including Usher syndrome, are characterized by initial loss of peripheral rod photoreceptors, leading to night blindness and progressive constriction of the visual field. The
time course of retinal degeneration in Usher syndrome is variable, but human children with USH1B show a reduction in the scotopic electroretinogram (ERGs), indicating a reduced function of
rod photoreceptors, as early as 6 months44,45,46, which is equivalent to 6 weeks in macaque infants. In contrast, in Mya, both rod and cone function appeared normal through 12 months of age.
The only suggestion of abnormal retinal development was a slight reduction in the thickness of the inner and outer segments of the photoreceptors in the central retina, as seen in macular
sdOCT scans. Current, generally available OCT imaging technology allows for measures of retinal thickness only in the central retina, not in the periphery where degeneration begins.
Therefore, without retinal histopathological analysis, we cannot exclude the possibility of subtle degenerative changes in the peripheral retina in the case of mosaic editing like Mya. It
remains to be seen if Mya will exhibit signs of retinitis pigmentosa later in life. However, the lack of a USH1B phenotype in Mya has clinical relevance as it indicates that maintaining or
achieving a population of ~ 50% of cells with functional MyoVIIA would suffice in treating individuals with USH1B. Also, the level of _MYO7A_ mutation in the sensory organs should be
determined in future studies, because normal hearing and vision could be the result of greater numbers of cells that lack the _MYO7A_ mutation in the eye and inner ear. MATERIALS AND METHODS
ETHICAL APPROVAL All protocols involving animals were approved by the ONPRC IACUC and conducted in accordance with the NIH Guidelines for the Care and Use of Laboratory Animals and ARRIVE
guidelines. General care and animal housing was previously described47,48. GRNA DESIGN Web-based algorithms (CRISPOR, Synthego, and IDT) were used to design 4 gRNAs targeting exon 3 of the
_MYO7A_ gene with optimal on-target efficiency and minimal off-target effects49. Hyb-gRNAs were purchased from Integrated DNA Technologies, and sgRNAs were synthesized and obtained from
Synthego. TARGET EFFICIENCY VALIDATION To confirm the targeting efficiency of the 4 gRNAs, _MYO7A_ target site amplicon was digested with Cas9 nuclease (New England Biolabs) and hyb-gRNAs. A
total of 100 ng of wild-type _MYO7A_ PCR amplicon was digested by 8 ng of Cas9 nuclease and four different concentrations (50, 10, 2, and 0.4 ng/μL) of hyb-gRNA. Reactions were incubated
for 30 min at 37 °C, 4 μg of RNAase was added to eliminate the hyb-gRNA, and samples were run on a 1.5% agarose gel. The CMMT cell line was used to test hyb-gRNA efficiencies alone or in
combination. At 75–80% confluency, 600 ng of Cas9 plasmid (Addgene, #64324) was transfected using Lipofectamine 2000 (Invitrogen), followed 24 h later with transfection of hyb-gRNAs using
RNAiMax (Invitrogen). A total of 6 pmol of hyb-gRNA was used for individual hyb-gRNA transfections while 3 pmol per hyb-gRNA was used for multi hyb-gRNA transfections. Genomic DNA was
obtained (QuickExtract, Epicentre) 2–3 days after transfection. The _MYO7A_ target region was amplified and subjected to a T7 endonuclease 1 (T7E1, New England Biolabs) assay according to
the manufacturer’s protocol using 200 ng of target amplicon or 150 ng target amplicon combined with 50 ng of wild-type amplicon were used because the inclusion of WT amplicon will ensure the
formation of dsDNA containing mismatches that result from Cas9 editing and easily discernable T7E1 cleavage products by gel electrophoresis. ZYGOTE INJECTION OF CRISPR/CAS9 Oocyte
collection and in vitro fertilization (IVF) were conducted as previously reported50. Zygotes were stripped free of sperm and transferred to warmed TALP-HEPES under oil. Cas9 nuclease and
sgRNAs were prepared as described26. Cas9 mRNA injection buffer contained 10 mM Tris–HCl, 10 mM NaCl, and 0.1 mM EDTA, whereas Cas9 protein injection buffer omitted the NaCl. Injection
materials included 45 ng/μL of hyb-gRNAs (15 ng/μL of each) with 100 ng/μL of Cas9 mRNA (Trilink) or 45 ng/μL sgRNAs (15 ng/μL of each) with 224 ng/μL of Cas9 nuclease (New England Biolabs).
Injections were into the cytoplasm and pronucleus when visible. Injected zygotes were then cultured (BO-IVC, IVF Bioscience) under oil in a 6%, 5%, 89% mixture of CO2, O2, and N2 at 37 °C
in humidified air, respectively. TE BIOPSY AND EMBRYO GENOTYPING An objective-mounted laser (Hamilton Thorne) was used to biopsy TE cells of blastocysts placed in warmed Biopsy Medium (IVF
Bioscience) under oil. The blastocyst was frozen using the global® Blastocyst Fast Freeze® Kit (LifeGlobal Group). Arrested embryos with the zona pellucida removed by acid Tyrode’s solution,
TE biopsy samples, and in some instances, biopsied blastocysts were subjected to WGA (Repli-g Single Cell, Qiagen). The resultant DNA was used to PCR amplify the _MYO7A_ target region and
amplicons were subjected to next-generation sequencing (NGS) using NEXTFLEX® Rapid DNA-Seq Kit (PerkinElmer). The DNA library was sequenced on an Illumina MiSeq platform and sequence reads
were assessed for indel mutations (Geneious software). OFF-TARGET EDITING ANALYSIS Putative off-target sites were identified using CAS-OFFinder51. Off-target regions were selected based on
exon regions over introns and intragenic regions with high homology at the seed sequence which is more permissive for off-target editing. Off-target sites were selected for analysis based on
the following parameters: they include exons, possess a high degree of homology to the seed sequence, and have less than 4 total mismatches between the gRNA and target sequences. Using
these criteria, three potential off-target sites for each gRNA target (Supplementary Table 2) were amplified by PCR. Resultant amplicons were subjected to adapter ligation and sequenced on
an Illumina MiSeq platform. CRISPresso2 software was used for sequence analysis52. EMBRYO TRANSFER Embryos selected for transfer were thawed (Global® Blastocyst Thawing Kit, Life Global) and
maintained for 2 h in 100 µL drops of BO-IVC under oil to monitor post-thaw viability. Embryos with blastocoel reformation or expansion were transferred into recipients as previously
described53. A transfer catheter was inserted into the oviduct and the embryo was deposited in 10 µL of TALP-HEPES. After embryo transfer, pregnancy was confirmed by ultrasound at
gestational day 28. _MYO7A_ GENOTYPING Genomic DNA was extracted from blood, buccal swabs, skin, and placental tissue and used to PCR amplify the targeted region of _MYO7A_ (primers detailed
in Supplementary Table 3). PCR amplicons were purified and prepared for NGS sequencing (NEXTFLEX® Rapid DNA-Seq Kit) using an Illumina MiSeq platform with roughly 5–10 thousand reads per
sample. Geneious software and the CRISPResso pipeline were used for sequence analysis. Single cell genotyping was conducted on leukocytes isolated from whole blood. Using a Becton Dickinson
Aria-II FACS, cells were sorted by morphology and then underwent WGA, with the resultant DNA being used for PCR amplification of _MYO7A_ exon 3. Purified amplicons were then Sanger
sequenced. Exon3 primers were used to detect large deletion on target locus and PCR amplicon was analyzed by Sanger sequencing. ASSESSMENT OF AUDITORY FUNCTION The animal was anesthetized
and placed within an IAC Acoustics MAC-3 RF-shielded Mini-Acoustical Chamber. The patency of the distal ear canal was confirmed by otoscopic evaluation. Middle ear function was tested at 1
kHz or 226 Hz by the ZODIAC Diagnostic Tympanometer (Model 8-04-16045). For 1-month and 3-month testing, stimuli were delivered in the ear canal via a custom acoustic assembly comprised of
two dynamic drivers (Etymotic Research ER4B-SG) and a miniature electret condenser microphone (Knowles FG-23329-P07). For testing at 6- and 12-months, stimuli were delivered with an open
field speaker (Fostex FF85WK) calibrated and monitored with a ¼-inch condenser microphone (Larson Davis Model 377C10). Acoustic stimuli were conducted as previously reported54. Five-ms tone
pip stimuli with 0.5-ms rise/fall times (cosine squared) in alternating polarity were presented in an interleaved stimulus train from 20 to 85 dB SPL in incremental steps of 5 dB at each
frequency tested. The order of frequencies was 26, 12, 4, 0.5, 16, 8 and 2 kHz presented at a stimulus rate of 60/s. The response from the electrodes was amplified 10,000 ×, filtered at 300
Hz–3 kHz band pass, and averaged for 300 samples at each frequency-level combination. Responses were recorded from subcutaneous electrodes located at the vertex, the ipsilateral mastoid, and
the shoulder. DPOAEs were recorded using the ear canal sound delivery apparatus controlled by Eaton Peabody Laboratory Cochlear Function Test Suite software (ver. 2.6.3.831). Primary f2
tones were presented at 2, 4, 12, 16 and 26 kHz with a primary frequency ratio f2/f1 of 1.2 and the f2 primary level 10 dB less than the f1 level. Primaries were incremented in 10 dB steps,
with f2 from 20 to 70 dB SPL. Ear-canal sound pressure was amplified, digitally sampled and averaged, Fast Fourier Transforms were computed and averaged, and the DPOAE and noise floor at
2f1–f2 were extracted. ASSESSMENT OF RETINAL STRUCTURE AND FUNCTION Retinal structure was evaluated under isoflurane anesthesia with retinal imaging, including color fundus photography and
fluorescein angiography (Zeiss FF450), sdOCT (Heidelberg Spectralis) and quantitative fundus autofluorescence55 (Spectralis Blue Peak mode), and ultra-widefield imaging (Optos
California)56,57,58,59. Retinal function was assessed with full-field electroretinography (Espion Diagnosys) under anesthesia with ketamine and xylazine. For each of these measures, results
for Mya were compared to those for age-matched normal rhesus infants. OCT imaging parameters included a standard scan (30° × 25°; ART:20; 61 lines with ~ 120 µm spacing), plus two high
density scans (#1:15° × 10°; ART:20; 49 lines with ~ 60 μm spacing; and #2: 15° × 5°; ART:20; 97 lines with ~ 30 μm spacing) for characterization of the macula/fovea and any salient features
of interest. The directional OCT method was performed as described to obtain an accurate measurement of the foveal outer nuclear layer60. Spectralis segmentation software (EyeExplorer
1.10.4) was used to obtain layer thicknesses, with manual inspection and corrections of each slice. Corneal curvature and axial length were measured with an IOL Master at each retinal
imaging session to allow conversion of retinal parameters from degrees of arc to mm. Ultra-wide field imaging in the peripheral retina, including pseudocolor, FAF, fluorescein angiography of
the retinal vasculature, and indocyanine angiography of the choroidal vasculature, was performed to identify peripheral changes associated with retinitis pigmentosa59. Full-field ERGs were
recorded with a Diagnosys Espion system using custom bipolar Burian–Allen contact lens electrodes and included photopic, flicker and scotopic intensity-response series, and a dark adaptation
series. STATISTICS ANALYSIS To compare the results of embryo development and target gene editing between the two conditions (mRNA and Protein), we conducted a two-tailed Fisher’s Exact
Test. A p value < 0.01 was considered statistically significant. REFERENCES * Boughman, J. A., Vernon, M. & Shaver, K. A. Usher syndrome: Definition and estimate of prevalence from
two high-risk populations. _J. Chronic Dis._ 36, 595–603. https://doi.org/10.1016/0021-9681(83)90147-9 (1983). Article CAS PubMed Google Scholar * Kimberling, W. J. _et al._ Frequency of
Usher syndrome in two pediatric populations: Implications for genetic screening of deaf and hard of hearing children. _Genet. Med._ 12, 512–516. https://doi.org/10.1097/GIM.0b013e3181e5afb8
(2010). Article CAS PubMed PubMed Central Google Scholar * Keats, B. J. & Corey, D. P. The usher syndromes. _Am. J. Med. Genet._ 89, 158–166 (1999). Article CAS Google Scholar *
Mathur, P. & Yang, J. Usher syndrome: Hearing loss, retinal degeneration and associated abnormalities. _Biochim. Biophys. Acta_ 1852, 406–420.
https://doi.org/10.1016/j.bbadis.2014.11.020 (2015). Article CAS PubMed Google Scholar * Kremer, H., van Wijk, E., Marker, T., Wolfrum, U. & Roepman, R. Usher syndrome: Molecular
links of pathogenesis, proteins and pathways. _Hum. Mol. Genet._ 15 SPEC NO 2, 262–270. https://doi.org/10.1093/hmg/ddl205 (2006). Article CAS Google Scholar * Colella, P. _et al._
Efficient gene delivery to the cone-enriched pig retina by dual AAV vectors. _Gene Ther._ 21, 450–456. https://doi.org/10.1038/gt.2014.8 (2014). Article CAS PubMed Google Scholar * Dyka,
F. M., Boye, S. L., Chiodo, V. A., Hauswirth, W. W. & Boye, S. E. Dual adeno-associated virus vectors result in efficient in vitro and in vivo expression of an oversized gene, MYO7A.
_Hum. Gene Ther. Methods_ 25, 166–177. https://doi.org/10.1089/hgtb.2013.212 (2014). Article CAS PubMed PubMed Central Google Scholar * Trapani, I. _et al._ Effective delivery of large
genes to the retina by dual AAV vectors. _EMBO Mol. Med._ 6, 194–211. https://doi.org/10.1002/emmm.201302948 (2014). Article CAS PubMed Google Scholar * Sachdeva, M. M. & Eliott, D.
Stem cell-based therapy for diseases of the retinal pigment epithelium: From bench to bedside. _Semin. Ophthalmol._ 31, 25–29. https://doi.org/10.3109/08820538.2015.1115253 (2016). Article
PubMed Google Scholar * Wilson, D. J. _et al._ Subretinal cell-based therapy: An analysis of surgical variables to increase cell survival. _Retina_ 37, 2162–2166.
https://doi.org/10.1097/IAE.0000000000001462 (2017). Article PubMed PubMed Central Google Scholar * Wang, L., Kempton, J. B. & Brigande, J. V. Gene therapy in mouse models of
deafness and balance dysfunction. _Front. Mol. Neurosci._ 11, 300. https://doi.org/10.3389/fnmol.2018.00300 (2018). Article CAS PubMed PubMed Central Google Scholar * Williams, D. S.
Usher syndrome: Animal models, retinal function of Usher proteins, and prospects for gene therapy. _Vis. Res._ 48, 433–441. https://doi.org/10.1016/j.visres.2007.08.015 (2008). Article CAS
PubMed Google Scholar * Sahly, I. _et al._ Localization of Usher 1 proteins to the photoreceptor calyceal processes, which are absent from mice. _J. Cell Biol._ 199, 381–399.
https://doi.org/10.1083/jcb.201202012 (2012). Article CAS PubMed PubMed Central Google Scholar * Mitalipov, S. M., Yeoman, R. R., Nusser, K. D. & Wolf, D. P. Rhesus monkey embryos
produced by nuclear transfer from embryonic blastomeres or somatic cells. _Biol. Reprod._ 66, 1367–1373. https://doi.org/10.1095/biolreprod66.5.1367 (2002). Article CAS PubMed Google
Scholar * Simerly, C. _et al._ Molecular correlates of primate nuclear transfer failures. _Science_ 300, 297. https://doi.org/10.1126/science.1082091 (2003). Article PubMed Google Scholar
* Ng, S. C. _et al._ The first cell cycle after transfer of somatic cell nuclei in a non-human primate. _Development_ 131, 2475–2484. https://doi.org/10.1242/dev.01118 (2004). Article CAS
PubMed Google Scholar * Mitalipov, S. M. _et al._ Reprogramming following somatic cell nuclear transfer in primates is dependent upon nuclear remodeling. _Hum. Reprod._ 22, 2232–2242.
https://doi.org/10.1093/humrep/dem136 (2007). Article CAS PubMed Google Scholar * NIH. _NHP Evaluation and Analysis Final Report—Part 2: Report of the Expert Panel on Challenges in
Assessing Nonhuman Primate Need and Resources for Biomedical Research._
https://orip.nih.gov/sites/default/files/NHP%20Evaluation%20and%20Analysis%20Final%20%20Report%20-%20Part%202%20Final%20508%2021Dec2018_002.pdf (2018). * Zuo, E. _et al._ One-step generation
of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs. _Cell Res._ 27, 933–945. https://doi.org/10.1038/cr.2017.81 (2017). Article CAS
PubMed PubMed Central Google Scholar * Weston, M. D. _et al._ Myosin VIIA mutation screening in 189 Usher syndrome type 1 patients. _Am. J. Hum. Genet._ 59, 1074–1083 (1996). CAS PubMed
PubMed Central Google Scholar * Jaijo, T. _et al._ MYO7A mutation screening in Usher syndrome type I patients from diverse origins. _J. Med. Genet._ 44, e71.
https://doi.org/10.1136/jmg.2006.045377 (2007). Article CAS PubMed PubMed Central Google Scholar * Hopper, K. J., Capozzi, D. K. & Newsome, J. T. Effects of maternal and infant
characteristics on birth weight and gestation length in a colony of rhesus macaques (_Macaca mulatta_). _Comp. Med._ 58, 597–603 (2008). CAS PubMed PubMed Central Google Scholar * Niu,
Y. _et al._ Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. _Cell_ 156, 836–843. https://doi.org/10.1016/j.cell.2014.01.027 (2014).
Article CAS PubMed Google Scholar * Hinz, J. M., Laughery, M. F. & Wyrick, J. J. Nucleosomes inhibit Cas9 endonuclease activity in vitro. _Biochemistry_ 54, 7063–7066.
https://doi.org/10.1021/acs.biochem.5b01108 (2015). Article CAS PubMed Google Scholar * Menoret, S. _et al._ Homology-directed repair in rodent zygotes using Cas9 and TALEN engineered
proteins. _Sci. Rep._ 5, 14410. https://doi.org/10.1038/srep14410 (2015). Article ADS CAS PubMed PubMed Central Google Scholar * Park, K. E. _et al._ Targeted gene knock-in by
CRISPR/Cas ribonucleoproteins in porcine zygotes. _Sci. Rep._ 7, 42458. https://doi.org/10.1038/srep42458 (2017). Article ADS CAS PubMed PubMed Central Google Scholar * Tang, L. _et
al._ CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein. _Mol. Genet. Genomics_ 292, 525–533. https://doi.org/10.1007/s00438-017-1299-z (2017). Article CAS PubMed
Google Scholar * Elaswad, A. _et al._ Effects of CRISPR/Cas9 dosage on TICAM1 and RBL gene mutation rate, embryonic development, hatchability and fry survival in channel catfish. _Sci.
Rep._ 8, 16499. https://doi.org/10.1038/s41598-018-34738-4 (2018). Article ADS CAS PubMed PubMed Central Google Scholar * Zuccaro, M. V. _et al._ Allele-specific chromosome removal
after Cas9 cleavage in human embryos. _Cell_ 183, 1650-1664 e1615. https://doi.org/10.1016/j.cell.2020.10.025 (2020). Article CAS PubMed Google Scholar * Kokkali, G. _et al._ Birth of a
healthy infant following trophectoderm biopsy from blastocysts for PGD of beta-thalassaemia major. _Hum. Reprod._ 20, 1855–1859. https://doi.org/10.1093/humrep/deh893 (2005). Article CAS
PubMed Google Scholar * Vilarino, M. _et al._ Mosaicism diminishes the value of pre-implantation embryo biopsies for detecting CRISPR/Cas9 induced mutations in sheep. _Transgenic Res._ 27,
525–537. https://doi.org/10.1007/s11248-018-0094-x (2018). Article CAS PubMed Google Scholar * Zhang, L. _et al._ Developmental potential of rhesus monkey embryos produced by in vitro
fertilization. _Biol. Reprod._ 51, 433–440. https://doi.org/10.1095/biolreprod51.3.433 (1994). Article CAS PubMed Google Scholar * Miller, J. C. _et al._ An improved zinc-finger nuclease
architecture for highly specific genome editing. _Nat. Biotechnol._ 25, 778–785. https://doi.org/10.1038/nbt1319 (2007). Article CAS PubMed Google Scholar * Szczepek, M. _et al._
Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. _Nat. Biotechnol._ 25, 786–793. https://doi.org/10.1038/nbt1317 (2007). Article CAS
PubMed Google Scholar * Cong, L. _et al._ Multiplex genome engineering using CRISPR/Cas systems. _Science_ 339, 819–823. https://doi.org/10.1126/science.1231143 (2013). Article ADS CAS
PubMed PubMed Central Google Scholar * Zhang, X. H., Tee, L. Y., Wang, X. G., Huang, Q. S. & Yang, S. H. Off-target effects in CRISPR/Cas9-mediated genome engineering. _Mol. Ther.
Nucleic Acids_ 4, e264. https://doi.org/10.1038/mtna.2015.37 (2015). Article CAS PubMed PubMed Central Google Scholar * Zheng, T. _et al._ Profiling single-guide RNA specificity reveals
a mismatch sensitive core sequence. _Sci. Rep._ 7, 40638. https://doi.org/10.1038/srep40638 (2017). Article ADS CAS PubMed PubMed Central Google Scholar * Carey, K. _et al._ Frequency
of off-targeting in genome edited pigs produced via direct injection of the CRISPR/Cas9 system into developing embryos. _BMC Biotechnol._ 19, 25. https://doi.org/10.1186/s12896-019-0517-7
(2019). Article PubMed PubMed Central Google Scholar * Salvi, R. _et al._ Inner hair cell loss disrupts hearing and cochlear function leading to sensory deprivation and enhanced central
auditory gain. _Front. Neurosci._ 10, 621. https://doi.org/10.3389/fnins.2016.00621 (2016). Article PubMed Google Scholar * Doyle, W. J., Saad, M. M. & Fria, T. J. Maturation of the
auditory brain stem response in rhesus monkeys (_Macaca mulatta_). _Electroencephalogr. Clin. Neurophysiol._ 56, 210–223. https://doi.org/10.1016/0013-4694(83)90075-5 (1983). Article CAS
PubMed Google Scholar * Ren, T. & Gillespie, P. G. Probing the cochlear amplifier by immobilizing molecular motors of sensory hair cells. _Neuron_ 76, 868–870.
https://doi.org/10.1016/j.neuron.2012.11.016 (2012). Article CAS PubMed Google Scholar * Ren, T., He, W. & Porsov, E. Localization of the cochlear amplifier in living sensitive ears.
_PLoS One_ 6, e20149. https://doi.org/10.1371/journal.pone.0020149 (2011). Article ADS CAS PubMed PubMed Central Google Scholar * Ruben, R. J. The developing concept of tonotopic
organization of the inner ear. _J. Assoc. Res. Otolaryngol._ 21, 1–20. https://doi.org/10.1007/s10162-019-00741-3 (2020). Article PubMed PubMed Central Google Scholar * Young, N. M.,
Mets, M. B. & Hain, T. C. Early diagnosis of Usher syndrome in infants and children. _Am. J. Otol._ 17, 30–34 (1996). CAS PubMed Google Scholar * Jacobson, S. G. _et al._ Retinal
disease course in Usher syndrome 1B due to MYO7A mutations. _Investig. Ophthalmol. Vis. Sci._ 52, 7924–7936. https://doi.org/10.1167/iovs.11-8313 (2011). Article CAS Google Scholar *
West, S. K. _et al._ Electroretinogram assessment of children with sensorineural hearing loss: Implications for screening. _J. AAPOS_ 19, 450–454.
https://doi.org/10.1016/j.jaapos.2015.08.001 (2015). Article PubMed Google Scholar * Peluffo, M. C., Barrett, S. L., Stouffer, R. L., Hennebold, J. D. & Zelinski, M. B. Cumulus-oocyte
complexes from small antral follicles during the early follicular phase of menstrual cycles in rhesus monkeys yield oocytes that reinitiate meiosis and fertilize in vitro. _Biol. Reprod._
83, 525–532. https://doi.org/10.1095/biolreprod.110.084418 (2010). Article CAS PubMed PubMed Central Google Scholar * Fayomi, A. P. _et al._ Autologous grafting of cryopreserved
prepubertal rhesus testis produces sperm and offspring. _Science_ 363, 1314–1319. https://doi.org/10.1126/science.aav2914 (2019). Article ADS CAS PubMed PubMed Central Google Scholar *
Concordet, J. P. & Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. _Nucleic Acids Res._ 46, W242–W245.
https://doi.org/10.1093/nar/gky354 (2018). Article CAS PubMed PubMed Central Google Scholar * Ramsey, C. & Hanna, C. In vitro culture of rhesus macaque (_Macaca_ _mulatta_) embryos.
_Methods Mol. Biol._ 2006, 341–353. https://doi.org/10.1007/978-1-4939-9566-0_23 (2019). Article CAS PubMed PubMed Central Google Scholar * Bae, S., Park, J. & Kim, J. S.
Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. _Bioinformatics_ 30, 1473–1475.
https://doi.org/10.1093/bioinformatics/btu048 (2014). Article CAS PubMed PubMed Central Google Scholar * Clement, K. _et al._ CRISPResso2 provides accurate and rapid genome editing
sequence analysis. _Nat. Biotechnol._ 37, 224–226. https://doi.org/10.1038/s41587-019-0032-3 (2019). Article CAS PubMed PubMed Central Google Scholar * Wolf, D. P. _et al._ In vitro
fertilization and embryo transfer in the rhesus monkey. _Biol. Reprod._ 41, 335–346. https://doi.org/10.1095/biolreprod41.2.335 (1989). Article CAS PubMed Google Scholar * Buran, B. N.
_et al._ Optimizing auditory brainstem response acquisition using interleaved frequencies. _J. Assoc. Res. Otolaryngol._ 21, 225–242. https://doi.org/10.1007/s10162-020-00754-3 (2020).
Article PubMed PubMed Central Google Scholar * McGill, T. J., Renner, L. M. & Neuringer, M. Elevated fundus autofluorescence in monkeys deficient in lutein, zeaxanthin, and omega-3
fatty acids. _Investig. Ophthalmol. Vis. Sci._ 57, 1361–1369. https://doi.org/10.1167/iovs.15-18596 (2016). Article Google Scholar * Fakin, A. _et al._ Fundus autofluorescence and optical
coherence tomography in relation to visual function in Usher syndrome type 1 and 2. _Vis. Res._ 75, 60–70. https://doi.org/10.1016/j.visres.2012.08.017 (2012). Article PubMed Google
Scholar * Lenassi, E. _et al._ Natural history and retinal structure in patients with Usher syndrome type 1 owing to MYO7A mutation. _Ophthalmology_ 121, 580–587.
https://doi.org/10.1016/j.ophtha.2013.09.017 (2014). Article PubMed Google Scholar * Yung, M., Klufas, M. A. & Sarraf, D. Clinical applications of fundus autofluorescence in retinal
disease. _Int. J. Retina Vitreous_ 2, 12. https://doi.org/10.1186/s40942-016-0035-x (2016). Article PubMed PubMed Central Google Scholar * Pradeep, S. & Prasad, D. S. _Atlas of
Wide-Field Imaging of Retinal Dystrophies_ (Springer, 2016). Google Scholar * Lujan, B. J. _et al._ Directional optical coherence tomography provides accurate outer nuclear layer and Henle
fiber layer measurements. _Retina_ 35, 1511–1520. https://doi.org/10.1097/IAE.0000000000000527 (2015). Article PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS
This work was supported by the NIH grant P51 OD011092 and a grant to M.N. from the Foundation Fighting Blindness. The collection of gametes, in vitro fertilization, embryo manipulation, and
embryo culture was made possible through the services provided by the ONPRC Assisted Reproductive Technologies (ART) Core and the Endocrine Technologies Core (ETC), which are supported by
P51 OD011092. Auditory assessments were supported by NIH grant R21 DC018126. The authors thank the excellent animal care and support by ONPRC Division of Animal Research and Resource Support
(ARRS) staff. AUTHOR INFORMATION Author notes * John P. Statz Present address: Division of Biological Sciences, University of Montana, Missoula, MT, 59812, USA * William Chan Present
address: University of Texas Southwestern Medical School, 5323 Harry Hines Blvd, Dallas, TX, 75390, USA * These authors contributed equally: Junghyun Ryu and John P. Statz. AUTHORS AND
AFFILIATIONS * Division of Reproductive and Developmental Sciences, Oregon National Primate Research Center, Oregon Health and Science University, Beaverton, OR, 97006, USA Junghyun Ryu,
John P. Statz, William Chan, Carol B. Hanna & Jon D. Hennebold * Assisted Reproductive Technologies Core, Oregon National Primate Research Center, Oregon Health and Science University,
Beaverton, OR, 97006, USA Fernanda C. Burch & Carol B. Hanna * Department of Otolaryngology, Oregon Hearing Research Center, Oregon Health and Science University, Portland, OR, 97239,
USA John V. Brigande, Beth Kempton & Edward V. Porsov * Division of Neuroscience, Oregon National Primate Research Center, Oregon Health and Science University, Beaverton, OR, 97006, USA
Lauren Renner, Trevor McGill & Martha Neuringer * Department of Ophthalmology, Casey Eye Institute, Oregon Health and Science University, Beaverton, OR, 97006, USA Trevor McGill &
Martha Neuringer * Vaccine and Gene Therapy Institute, Oregon Health and Science University, Beaverton, OR, 97006, USA Benjamin J. Burwitz * Department of Obstetrics and Gynecology, Oregon
Health and Science University, Portland, OR, 97239, USA Jon D. Hennebold Authors * Junghyun Ryu View author publications You can also search for this author inPubMed Google Scholar * John P.
Statz View author publications You can also search for this author inPubMed Google Scholar * William Chan View author publications You can also search for this author inPubMed Google
Scholar * Fernanda C. Burch View author publications You can also search for this author inPubMed Google Scholar * John V. Brigande View author publications You can also search for this
author inPubMed Google Scholar * Beth Kempton View author publications You can also search for this author inPubMed Google Scholar * Edward V. Porsov View author publications You can also
search for this author inPubMed Google Scholar * Lauren Renner View author publications You can also search for this author inPubMed Google Scholar * Trevor McGill View author publications
You can also search for this author inPubMed Google Scholar * Benjamin J. Burwitz View author publications You can also search for this author inPubMed Google Scholar * Carol B. Hanna View
author publications You can also search for this author inPubMed Google Scholar * Martha Neuringer View author publications You can also search for this author inPubMed Google Scholar * Jon
D. Hennebold View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.N. and J.D.H. designed and supervised this project. J.R. and J.P.S.
performed the gene editing experiments and analyzed data. W.C. conducted genotyping experiments. C.R., F.M.B., and C.B.H. performed in vitro fertilization, embryo micromanipulation, and
embryo culture. J.V.B., B.K., E.P., L.R., M.N., and T.M. conducted phenotyping of Mya. J.R and J.P.S. wrote the manuscript. All authors helped with the writing and editing of the manuscript.
CORRESPONDING AUTHOR Correspondence to Jon D. Hennebold. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION. RIGHTS AND
PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any
medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The
images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is
not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission
directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Ryu, J.,
Statz, J.P., Chan, W. _et al._ CRISPR/Cas9 editing of the MYO7A gene in rhesus macaque embryos to generate a primate model of Usher syndrome type 1B. _Sci Rep_ 12, 10036 (2022).
https://doi.org/10.1038/s41598-022-13689-x Download citation * Received: 11 March 2022 * Accepted: 26 May 2022 * Published: 16 June 2022 * DOI: https://doi.org/10.1038/s41598-022-13689-x
SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to
clipboard Provided by the Springer Nature SharedIt content-sharing initiative