Evaluating the performance characteristics of five lateral flow assays for the detection of the sars-cov-2 nucleocapsid antigen
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT In response to the COVID-19 pandemic, lateral flow assays (LFAs) for the detection of SARS-CoV-2 antigen have been proposed as a complementary option to the more costly and time
consuming reverse-transcriptase polymerase chain reaction (RT-PCR). We assessed five commercially available SARS-CoV-2 antigen detecting LFAs (ASSUT EUROPE (Rome, Italy), Besthree (Taizhou,
China), Encode (Zhuhai, China), Fortress (Antrim UK), and Hughes Medical (Buckinghamshire, UK), using samples collected from hospitalised individuals with COVID-19 and compared these results
against established RT-PCR assays with the aim of estimating test performance characteristics. We performed a diagnostic accuracy study of the five LFAs on 110 inpatients with confirmed
COVID-19 and 75 COVID-19 negative control participants. Assay evaluation was performed using a modified version of each manufacturer’s protocol allowing for parallel testing of a single
sample on multiple assays. Additional variables were studied including infection acquisition, oxygenation requirements at time of swabbing, and patient outcomes. The 110 patients were 48%
(53) female, with mean age 67 years (range 26–100 years), and 77% (85) cases were community onset SARS-CoV-2. Across the five assays, sensitivity ranged from 64 (95% CI 53–73) to 76% (95% CI
65–85); Fortress performed best with sensitivity of 76% (95% CI 65–85). Specificity was high across all assays with 4/5 LFAs achieving 100%. LFA sensitivity was not dependant on RT-PCR
cycle thresholds. SARS-CoV-2 antigen detecting LFAs may complement RT-PCR testing to facilitate early diagnosis and provide community testing strategies for identification of patients with
COVID-19, however we find suboptimal test performance characteristics across a range of commercially available manufacturers, below WHO and MHRA pre-set sensitivity performance thresholds.
With such variation in sensitivity between LFAs and PCR testing and between assay brands, we advise caution in the deployment of LFAs outside of environments with clinical oversight. SIMILAR
CONTENT BEING VIEWED BY OTHERS EVALUATION OF THE ACCESS BIO CARESTART RAPID SARS-COV-2 ANTIGEN TEST IN ASYMPTOMATIC INDIVIDUALS TESTED AT A COMMUNITY MASS-TESTING PROGRAM IN WESTERN
MASSACHUSETTS Article Open access 09 December 2022 FASTER DETECTION OF ASYMPTOMATIC COVID-19 CASES AMONG CARE HOME STAFF IN ENGLAND THROUGH THE COMBINATION OF SARS-COV-2 TESTING TECHNOLOGIES
Article Open access 29 March 2024 POINT-OF-CARE SARS-COV-2 SEROLOGICAL ASSAYS FOR ENHANCED CASE FINDING IN A UK INPATIENT POPULATION Article Open access 12 March 2021 INTRODUCTION In
response to the COVID-19 pandemic, testing capacity for clinical diagnostics and for screening have been stretched at a global level. Increasing demand has meant that novel testing platforms
and protocols are required, with an expectation that they will play a role in both individual healthcare and community testing programmes, national population studies, and international
epidemiological guidance. Reverse transcriptase polymerase chain reaction (RT-PCR) is considered the gold standard test for the detection of SARS-CoV-2 infection, but has a significant
processing time requiring specific laboratory infrastructure1. Whilst some advances have been made in laboratory-free RT-PCR platforms, these are still not in widespread use in large scale
community testing2,3. A faster and more portable alternative is required and lateral flow assays (LFAs) for the detection of SARS-COV-2 antigen have been proposed as a complementary option4.
During the rapid development of SARS-CoV-2 antibody lateral flow assays, significant inter-manufacturer variation was seen, with some performing particularly poorly5,6,7. If antigen LFAs
are to be used to complement PCR testing, evaluation for any similar inter-manufacturer variation must be analysed. Whilst laboratory based assessment of these rapid antigen LFA assays has
occurred, evaluation of assays in the setting of intended use with both sample and operator skill-sets who are representative of clinical and community testing sites remains unstudied. In
the UK, testing has focused on one specific manufacturer, procured at a national level8,9, whilst competitor assays also remain unstudied. In this analysis we assessed five commercially
available SARS-CoV-2 antigen LFAs using samples collected from hospitalised individuals with COVID-19 and compared these results against established RT-PCR assays with the aim of estimating
test performance characteristics. METHODS STUDY DESIGN We performed a prospective diagnostic accuracy study, independent of manufacturers, to evaluate five commercially available SARS-CoV-2
antigen point-of-care lateral flow assays between November 2020 and February 2021. This was a single centre study, based in a large urban hospital, comprising 110 in-patients and 75
SARS-CoV-2 negative controls. Clinicians identified participants, collected clinical details including the oxygen requirements and infection acquisition source of each patient, and performed
nasopharyngeal swabs for inoculation of the lateral flow assays. Follow-up data on patient survival was also collected at the end of the study. SARS-CoV-2 antigen LFAs were identified
through our procurement team as viable alternatives to the nationally procured LFA8,9. Initial assessment was performed with all assays required to meet the following criteria: a
cassette-based design with visual read-out result in less than 20 min, and manufacturer reported sensitivity of > 95% and specificity of > 95%, availability for purchase in the United
Kingdom, and accreditation via a CE mark. Five assays were selected for in-depth evaluation; ASSUT EUROPE (Rome, Italy), Besthree (Taizhou, China), Encode (Zhuhai, China), Fortress (Antrim
UK), and Hughes Medical (Buckinghamshire, UK). The target of all selected assays was the SARS-CoV-2 nucleocapsid protein antigen. All evaluated assays were approved for nasopharyngeal
sampling. As all assays were CE marked, this service evaluation was undertaken as a validation of these in vitro diagnostic devices in line with the UK Standards for Microbiological
Investigation structure10. The service evaluation was registered with the hospital Clinical Governance Department, and all participants provided informed consent for use of their samples for
the purpose of assay validation. SARS-COV-2 POSITIVE COHORT Patients confirmed to have COVID-19 were identified via a centralised daily report of all SARS-CoV-2 results performed in the
previous 24 h within our Trust. Patients’ electronic health records were examined to assess for suitability according to the inclusion criteria; current inpatient at Chelsea and Westminster
Hospital NHS Foundation Trust, positive RT-PCR test for SARS-CoV-2 within the previous 72 h, symptom onset < 14 days (in those with symptoms), ability to provide informed consent.
Patients were excluded if; age < 18 years old, unable to consent due to pre-existing medical condition or acute alteration in an individual’s condition, symptom onset > 14 days, PCR
positive for SARS-CoV-2 > 72 h, asymptomatic without a negative swab within the previous 14 days. The laboratory serving the hospital network used a variety of PCR platforms to derive
COVID-19 status, including AusDiagnostics (Mascot, Australia), Roche Cobas 6600 (Roche Molecular Systems, New Jersey, US), and Abbott RealTime (Illinois, US) platforms. SARS-COV-2 NEGATIVE
CONTROL COHORT Negative controls were recruited from hospital staff, and students from Imperial College School of Medicine. In line with NHS England guided testing of healthcare
professionals, all individuals recruited undertook routine twice weekly LFA testing using the aforementioned nationally procured Innova assay. Additionally, all recruited members from the
Haematology, Oncology and HIV teams were undergoing weekly PCR testing in keeping with local policy on health care professionals working with immunocompromised patients. All individuals
recruited as negative controls were asymptomatic at time of testing and had a negative Innova LFA or RT-PCR within the previous 72 h. EVALUATION PROTOCOL Assay evaluation was performed using
a modified version of each manufacturer’s protocol. This allowed for parallel testing of a single sample (nasopharyngeal swab) across the five assays. These modifications included an
increase in the quantity of extraction buffer used from 300 to 600 µl and the use of a single extraction solution with sodium azide 0.09% preservative. This protocol deviation was evaluated
using five positive and five negative _ACON Biotech5_ (Hangzhou, China) SARS-CoV-2 positive control swabs on each of the five assays and compared to the original manufacturers protocol.
There was 100% agreement between the modified and the original protocol in positive and negative control cases. The nasopharyngeal swabbing procedure followed manufacturer protocol and all
samples were taken by a trained member of the research team to ensure consistent sampling technique. Samples were stored in refrigerated temperatures (2–4 degrees centrigrade). All assays
with a visible control line were deemed valid. Chromatographic results on each assay were read by a single member of staff trained in the use of lateral flow assays including interpretation
of results. In the case of an inconclusive result, a second member of the research team was consulted to reach a final decision. STATISTICAL ANALYSIS Each LFA result was compared to the
RT-PCR result. Cycle threshold values were not available for all RT-PCR samples and thus absent results were imputed via multiple imputations using chained equations11. This technique
facilitates the use of missing data whilst propagating the uncertainty in estimating missing data, thus minimising selection bias. The dataset is imputed 20 times, using differing random
seeds, resulting in 20 different possible values for each missing value. Each imputed dataset’s performance is then aggregated, and the spread is calculated and displayed for the appropriate
metric. To understand the effects of the RT-PCR cycle threshold (CT) and temporal delay between swabbing on the accuracy of LFAs, receiver operating characteristic (ROC) curves were
constructed for each assay. ETHICAL APPROVAL This evaluation was commissioned as a service evaluation by the COVID Testing Committee of Chelsea & Westminster NHS Foundation Trust, on 20
October 2020. The study was reviewed by the Chelsea & Westminster NHS Foundation Trust Research and Development Office and deemed a verification of a CE marked in vitro diagnostic test;
therefore, informed consent was required to participate but this was not required to be in written format. Aggregated data was analysed under the Health Service Control of Patient
Information Regulations (2002) general notice that patient data for a COVID-19 purposes may be used for research as stated by the UK Secretary of State for Health and Social Care. The study
was conducted in accordance with relevant guidelines and regulations including the Declaration of Helsinki. RESULTS PATIENT/NEGATIVE CONTROL COHORT As detailed, the SARS-CoV-2 positive
cohort consisted of 110 inpatients, with each assay tested against 75 of these individuals. The cohort was almost evenly split by gender, 52% (57) male and 48% (53) female and ages ranged
from 26 to 100 years old, with a mean age of 67 years. Almost half (46%, 51) of the cohort were in the higher risk age category (> 70 years old). Community acquisition of SARS-CoV-2
accounted for 77% (85) of our positive cohort with the remaining 23% (25) resulting from nosocomial transmission (see Table 1). Hypoxia requiring supplemental oxygen was a common symptomatic
feature. At the point of LFA sampling, 50% (55) of patients were not requiring supplemental oxygen, 38% (42) were receiving oxygen via nasal cannula or by non-rebreathe and Venturi face
masks, while 12% (13) were receiving positive pressure non-invasive ventilation. At the completion of data collection 61% (67) of the cohort had been discharged from hospital, 15% (17)
remained in hospital undergoing rehabilitation, and 24% (26) of the cohort had died (see Table 1). In total, 75 negative control individuals were enrolled in the validation process. The
cohort was 51% (38) male and 49% (37) female with an age range of 21–64 years (mean: 34 years of age) (see Table 2). LFA TEST PERFORMANCE CHARACTERISTICS The sensitivity of the LFAs ranged
from 64% (95% CI 53–73) to 76% (95% CI 65–85). Of the assays tested, Fortress performed best with a sensitivity of 76% (95% CI 65–85). Specificity based on testing SARS-CoV-2 volunteers was
100% (95% CI 95–100) for four of the LFA’s), except for Encode with 99% (95% CI 92–100) (see Table 3, Supplementary Receiver Operating Curve Figures). IMPACT ON TIME FROM RT-PCR TO LFA TEST
PERFORMANCE CHARACTERISTICS Time from RT-PCR swab to LFA swab was examined with 46% (51) of participants sampled within 24 h, 34% (38) within 48 h and 20% (21) within 72 h of a positive
RT-PCR test. No patients were sampled at greater than 72 h of a confirmed RT-PCR result. None of the LFAs showed significant dependence on the time between RT-PCR tests and LFA swab.
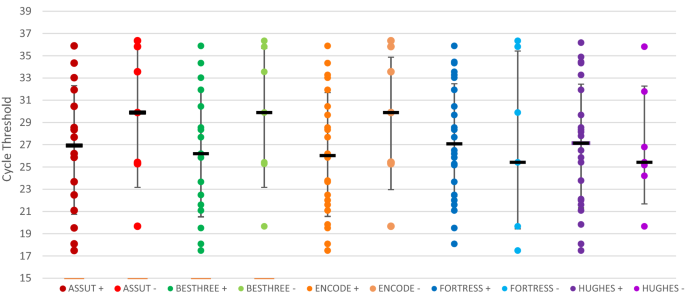
CORRELATION OF RT-PCR CYCLE THRESHOLD AND LFA TEST PERFORMANCE CHARACTERISTICS The sensitivity of each LFA was explored as a surrogate marker of the RT-PCR cycle threshold using ROC curves.
No particular test demonstrated reliance on a specific RT-PCR cycle threshold to achieve positivity (Fig. 1). Instead, across all five assays there were relatively constant median cycle
thresholds (of around 27) among those LFDs that were positive, and among those LFDs that were negative, suggesting that likelihood of nasopharyngeal antigen detection is independent of the
PCR cycle threshold. DISCUSSION Our study finds variation between 5 major manufacturer’s SARS-CoV-2 antigen lateral flow devices, in particular there is marked variation in sensitivity when
compared to RT-PCR. While other studies examining antigen detecting LFAs have utilised recombinant nucleoproteins, viral culture samples and stored respiratory samples, our study
demonstrates significant inter-manufacturer sensitivity (and particularly variability in time-from-RT-PCR-positive) using clinical in situ methodology. The range of manufacturer reported
sensitivity values (78% to 99%) differed substantially from the sensitivity determined in our evaluation (64% to 76%, 95% CI (53–85)), highlighting the importance of independent assessment
of product performance in real clinical settings with representative clinician-user skill sets. Our findings are corroborated by results from a PHE Porton Down study which showed a drop in
sensitivity of the UK government procured Innova antigen LFA8,12 when real-world samples, and operators, are used. The between-assay variation in sensitivity we demonstrated also suggests
that interchangeability between different manufacturers could significantly impact accuracy in testing. Based on our evaluation, none of the five assays meet the WHO and MHRA’s priority
target product profiles for COVID‐19 diagnostics, where acceptable sensitivity is deemed ≥ 80% and specificity ≥ 97%. Thus, none can be considered as a replacement for laboratory-based
RT-PCR13,14. They may, however, represent a useful diagnostic adjunct in the current SARS-CoV-2 pandemic, its potential future endemic presence, and in regions where RT-PCR testing remains
limited. With any highly infectious condition, time sensitive decisions regarding patient placement in hospital are required and laboratory-based RT-PCR cannot always meet this demand.
SARS-CoV-2 antigen LFAs may be best suited to situations where confirmatory RT-PCR testing can be performed once the patient is in a more secure environment. There are several limitations to
our study including deviation from the manufacturer protocol, although our verification with set laboratory controls indicated this had no effect. Secondly, we undertook non-contemporaneous
RT-PCR and LFA sampling. This decision was pragmatic, as unlike antibody LFAs which can be verified using excess/waste serum, antigen LFAs require nasal or nasopharyngeal samples for which
there is no excess. Tolerability of repeated nasopharyngeal swabs impacts the ability to perform head-to-head comparisons on individual patients. Variation in quantity and quality of
sampling compared to the initial RT-PCR swabs may impact on the reported assay sensitivity, but we mitigated this by using clinicians experienced in nasopharyngeal swabbing, to obtain all
samples. There remains a possibility that an individual’s antigen positive status may have waned after their initial RT-PCR positive result, but our analysis of the impact of
time-between-sampling demonstrated minimal impact of this variable, implying that this is unlikely to have had a significant impact on the LFA poor sensitivity. Finally, RT-PCR testing was
performed on a number validated platforms, and CT was not available to enable comparison for all assays (some were tested on the dnaNudge platform)2. Reassuringly, our investigation of
correlation between RT-PCR CT and LFA positivity demonstrated little effect here, yet in other centres there has been a demonstrable correlation between the likelihood of LFD positivity and
RT-PCR cycle thresholds13. We would caution relating this to infectivity however, given yet other groups have demonstrated that RT-PCR cyle thresholds do not necessarily correlate to
infectivity14. In conclusion, while SARS-CoV-2 antigen LFAs may complement RT-PCR testing to facilitate early diagnosis and provide community testing strategies for identification of
patients with COVID-19, we find suboptimal test performance characteristics across a range of commercially available manufacturers, below pre-set sensitivity performance thresholds15,16.
With limited access to healthcare facilities due to pandemic restrictions, the ability for patients to perform home sampling greatly reduces the demand on hospital services and the need for
unecessary travel to, and footfall within, clinical settings. Demand for a rapid, self-administered point of care diagnostic tool remains paramount, however with such variation in
sensitivity between LFAs and RT-PCR testing and between assay brands, we advise caution in the deployment of LFAs outside of environments with clinical oversight. DATA AVAILABILITY The data
analysed during the current study and further details on the assays are available from the corresponding author (LSPM; [email protected]) on reasonable request, as long as this meets
local ethical and research governance criteria. REFERENCES * WHO. _Establishment of PCR in Developing Countries_ https://apps.who.int/iris/bitstream/handle/10665/205020/B4745.pdf?sequence=1.
Accessed 24 Mar 2022 (2011). * Gibani, M. M. _et al._ Assessing a novel, lab-free, point-of-care test for SARS-CoV-2 (CovidNudge): A diagnostic accuracy study. _Lancet Microbe_ 1(7),
e300–e307 (2020). Article CAS Google Scholar * Moore, L. S. P. Near-patient SARS-CoV-2 molecular platforms: New-old tools for new-old problems. _Lancet Respir. Med._ 8(12), 1161–1163
(2020). Article CAS Google Scholar * Dinnes, J. _et al._ Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. _Cochrane Database Syst. Rev._ 8,
CD013705 (2020). PubMed Google Scholar * Pallett, S. J. C. _et al._ Point-of-care serological assays for delayed SARS-CoV-2 case identification among health-care workers in the UK: A
prospective multicentre cohort study. _Lancet Respir. Med._ 8(9), 885–894 (2020). Article CAS Google Scholar * Moshe, M. _et al._ SARS-CoV-2 lateral flow assays for possible use in
national covid-19 seroprevalence surveys (React 2): Diagnostic accuracy study. _BMJ_ 372, n423 (2021). Article Google Scholar * Flower, B. _et al._ Clinical and laboratory evaluation of
SARS-CoV-2 lateral flow assays for use in a national COVID-19 seroprevalence survey. _Thorax_ 75(12), 1082–1088 (2020). Article Google Scholar * University, P. O. _Preliminary Report from
the Joint PHE Porton Down & University of Oxford SARS-CoV-2 Test Development and Validation Cell: Rapid Evaluation of Lateral Flow Viral Antigen Detection Devices (LFDs) for Mass
Community Testing_ (2020). * Mahase, E. Covid-19: Innova lateral flow test is not fit for “test and release” strategy, say experts. _BMJ_ https://doi.org/10.1136/bmj.m4469 (2020). Article
PubMed Google Scholar * PHE. _UK Standards for Microbiology Investigations Q1: Evaluations, Validations and Verifications of Diagnostic Tests_.
https://www.gov.uk/government/publications/smi-q-1-commercial-and-in-house-diagnostic-tests-evaluations-and-validations2017. * White, I. R., Royston, P. & Wood, A. M. Multiple imputation
using chained equations: Issues and guidance for practice. _Stat. Med._ 30(4), 377–399 (2011). Article MathSciNet Google Scholar * Iacobucci, G. Covid-19: Mass population testing is
rolled out in Liverpool. _BMJ_ 371, m4268 (2020). Article Google Scholar * Pickering, S. _et al._ Comparative performance of SARS-CoV-2 lateral flow antigen tests and association with
detection of infectious virus in clinical specimens: A single-centre laboratory evaluation study. _Lancet Microbe_ 2(9), e461–e471 (2021). Article CAS Google Scholar * Singanayagam, A.
_et al._ Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. _Euro Surveill._ 25(32), 2001483 (2020). Article
CAS Google Scholar * WHO. _Target Product Profiles for Priority Diagnostics to Support Response to the COVID-19 Pandemic v.1.0_.
https://cdn.who.int/media/docs/default-source/blue-print/who-rd-blueprint-diagnostics-tpp-final-v1-0-28-09-jc-ppc-final-cmp92616a80172344e4be0edf315b582021.pdf?sfvrsn=e3747f20_1&download=true.
Accessed 24 Mar 2022 (2020). * MHRA. _Target Product Profile: Point of Care SARS-CoV-2 Detection Tests_.
https://www.gov.uk/government/publications/how-tests-and-testing-kits-for-coronavirus-covid-19-work/target-product-profile-point-of-care-sars-cov-2-detection-tests. Accessed 24 Mar 2022
(2020). Download references FUNDING JH received funding in the form of a fellowship from the Joint Research Committee of CW+ and the Westminster Medical School Research Trust. AUTHOR
INFORMATION AUTHORS AND AFFILIATIONS * Chelsea and Westminster NHS Foundation Trust, 369 Fulham Road, London, SW10 9NH, UK J. Heskin, A. Al-Hindawi, G. W. Davies, M. Rayment, N. Mughal, R.
Jones & L. S. P. Moore * Centre of Defence Pathology, Royal Centre for Defence Medicine, Queen Elizabeth Hospital Birmingham, Mindelsohn Way, Edgbaston, Birmingham, B15 2WB, UK S. J. C.
Pallett * North West London Pathology, Imperial College Healthcare NHS Trust, Fulham Palace Road, London, W6 8RF, UK N. Mughal, P. Randell & L. S. P. Moore * NIHR Health Protection
Research Unit in Healthcare Associated Infections & Antimicrobial Resistance, Imperial College London, Du Cane Road, London, UK L. S. P. Moore Authors * J. Heskin View author
publications You can also search for this author inPubMed Google Scholar * S. J. C. Pallett View author publications You can also search for this author inPubMed Google Scholar * A.
Al-Hindawi View author publications You can also search for this author inPubMed Google Scholar * G. W. Davies View author publications You can also search for this author inPubMed Google
Scholar * M. Rayment View author publications You can also search for this author inPubMed Google Scholar * N. Mughal View author publications You can also search for this author inPubMed
Google Scholar * P. Randell View author publications You can also search for this author inPubMed Google Scholar * R. Jones View author publications You can also search for this author
inPubMed Google Scholar * L. S. P. Moore View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS All listed authors made substantial contributions
to the conception or design of the work; or to the acquisition and analysis of data for the work; and drafting the work or revising it critically ahead of submission for publication. The
corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. J.H.—data curation, formal analysis, investigation,
writing—original draft (including figures); S.J.C.P.—methodology, data curation, writing—review and editing; A.A.H.—data analysis, statistical methodology, writing; N.M.—conceptualization,
supervision; P.R.—conceptualization, data curation, laboratory analysis, writing; R.J.—conceptualization, writing—review and editing; G.W.D.—conceptualization, supervision, project
administration, writing—review and editing; M.R.—conceptualization, data curation, writing—review and editing; L.S.P.M.—conceptualization, supervision, writing—review and editing, project
administration. CORRESPONDING AUTHOR Correspondence to J. Heskin. ETHICS DECLARATIONS COMPETING INTERESTS JH received research funding from CW+ Charity and the Westminster Medical School
Research Trust and received honoraria from Gilead (2020). SJCP has received a research Grant from the Scientific Exploration Society/Viscount Gough, outside the submitted work. NM has
received speaker fees from Beyer (2016) and Pfizer (2019) and received educational support from Eumedica (2016) and Baxter (2017). LSPM has consulted for or received speaker fees from
bioMerieux (2013–2022), Pfizer (2018–2022), Eumedica (2016–2022), DNAelectronics (2015–2018), Dairy Crest (2017–2018), Umovis Lab (2020–2021), Shionogi (2021–2022), Kent Pharma (2021),
Sumitovant (2021–2022), and Pulmocide (2021), and received research grants from the National Institute for Health Research (2013–2019), CW+ Charity (2018–2022) and LifeArc (2020–2022). RJ
has received honoraria, speaker fees, travel support and/or research grant funding from Gilead, ViiV Healthcare, BMS, Abbvie, Janssen and Merck. All other authors declare no competing
interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY FIGURE S1. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing,
adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons
licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise
in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the
permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Heskin, J., Pallett, S.J.C., Al-Hindawi, A. _et al._ Evaluating the performance characteristics of five lateral flow assays for the detection
of the SARS-CoV-2 nucleocapsid antigen. _Sci Rep_ 12, 8811 (2022). https://doi.org/10.1038/s41598-022-12805-1 Download citation * Received: 30 September 2021 * Accepted: 11 May 2022 *
Published: 25 May 2022 * DOI: https://doi.org/10.1038/s41598-022-12805-1 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link
Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative