New male specific markers for hop and application in breeding program
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Male specific DNA sequences were selected from a Diversity Arrays Technology (DArT) mapping study to evaluate their suitability for determination of the sex phenotype among young
seedlings in a hop (_Humulus lupulus_ L.) breeding program. Ten male specific DArT markers showed complete linkage with male sex phenotype in three crossing families. Following optimization,
four were successfully converted into PCR markers and a multiplex PCR approach for their use was developed. Among 197 plants (97 from the world collection; 100 from three segregating
families), 94–100% positive correlation with sex phenotypic data was achieved for the single PCR amplification, whereas the multiplex approach showed 100% correlation. To develop a fast and
low-cost method, crude sample multiplex PCR was evaluated in 253 progenies from 14 segregating populations without losing accuracy. The study describes, for the first time, the routine
application of molecular markers linked to male sex in an intensive Slovenian hop breeding program. The methods described could be employed for screening of sex at the seedling stage in
other hop programs worldwide, thereby saving resources for desirable female plants. SIMILAR CONTENT BEING VIEWED BY OTHERS CONSTRUCTION OF A BREEDING PARENT POPULATION OF _POPULUS TOMENTOSA_
BASED ON SSR GENETIC DISTANCE ANALYSIS Article Open access 29 October 2020 MAP AND SEQUENCE-BASED CHROMOSOME WALKING TOWARDS CLONING OF THE MALE FERTILITY RESTORATION GENE _RF5_ LINKED TO
_R__11_ IN SUNFLOWER Article Open access 12 January 2021 CHROMOSOME GENOMICS FACILITATES THE MARKER DEVELOPMENT AND SELECTION OF WHEAT-_AEGILOPS BIUNCIALIS_ ADDITION, SUBSTITUTION AND
TRANSLOCATION LINES Article Open access 22 November 2023 INTRODUCTION Hop (_Humulus lupulus_) is a perennial, dioecious, wind-pollinated species. The lateral shoots of the female hop plant
carry the cones that are essential to the beer brewing industry. Plant secondary metabolites that accumulate in hop cones contribute to bitterness (bitter acids), aroma (essential oils), and
stability in beer (polyphenols1,2), and exhibit pharmacological effects (anti-carcinogenic, anti-inflammatory and phytoestrogenic3). Global hop production occupies up to 60.000 ha4,
including acreage in Europe, USA, China, South Africa, Australia and New Zealand. Hop cultivars are clonally propagated for deployment, and as such may be highly locally adapted, with the
result that each hop growing jurisdiction tends to support a local hop breeding and cultivar development program. Hop breeding programs typically aim to increase yield (through plant
architecture or resistence/tolerance to abiotic or biotic stresses), and improve quality. Progress in hop breeding is limited by the biology of hop. Hop can take up to two years under field
conditions to reach reproductive maturity5,6. Furthermore, as the female hop cones are the commercial product it is difficult to relate the commercial requirements with the male phenotype
since cones do not develop on male plants. These factors, combined with high levels of heterozygosity, limit progress in hop breeding. Furthermore, the presence of male plants adjacent to
commercial hop fields or breeding populations results in the presence of seed in hop cones, which increases the weight of cones, but can reduce their brewing quality, and therefore their
value. Since the hop plant is wind pollinated, a single male plant in the hop field or its vicinity can cause broad scale damage to the crop7. In dioecious species such as hop, each
individual normally produces either male or female reproductive organs8. Dioecy has been observed in about 9% of the ~319.000 known plant species, with levels higher among bryophytes (62%)
and gymnosperms (36%), while in angiosperms only about 5% of species are dioecious9. In almost all studied dioecious species, the sex phenotype is linked to genetic differences between male
and female plants at least at one locus on a pair of chromosomes10. Renner and Ricklef11 also reported that dioecy is more common in wind-pollinated species in comparison to animal
pollinated plants. Molecular markers are a useful tool for determination of sex in dioecious plants before the phenotypic differences become detectable as the plants enter their reproductive
phase. Several reports on identification of sex-specific markers in other plants have been published, such as in rattan _Calamus guruba_ Buch.-Ham12, _Momordica dioica_13, _Coccinia
grandis_14, _Carica papaya_15, and _Cannabis sativa_16. Hop (_H. lupulus_ var. _lupulus_) is normally diploid (2n = 2x = 20) with nine autosomal bivalents and two heteromorphic sex
chromosomes. Female plants have a pair of X chromosomes, while male plants have an XY pair with the Y chromosome being smaller8,17,18,19. In all other species of the genus _Humulus_, such as
_H. lupulus_ var. _cordifolius_, _H. japonicus_, _H. yunnanensis_ and _H. acetosa_, the sex is expressed based on a multi-sex chromosome system20,21,22. Expression of sex phenotype in hop
is also influenced by the X: autosomes ratio8, indicating that the genes controlling pollen development are in the sex determining region and structural genes located on autosomes17,21. In
hop, monoecious plants occasionally occur, especially among progeny of specific crosses. Škof _et al_.23 revealed that a high percentage of monoecious hop plants were triploids. Experimental
work of this study focusses on diploid germplasm. Since the hop karyotype contains sex chromosome Y, which is transmitted from generation to generation only by the male line, it was first
assumed that majority of polymorphisms on Y chromosome should be male specific. Polley _et al_.6, on the basis of one cross, isolated a male molecular marker linked to sequences on the Y
chromosome. Čerenak and Javornik24 tested the published marker in the Slovenian breeding program, but the marker did not multiply in diploid and tetraploid forms of the Japanese male hop
No3-38, and therefore, there was an assumption that the marker developed was specific to European hops. Patzak _et al_.5 further tested its suitability in four families from the Czech
breeding program and they also observed that the marker was not absolutely linked to the male phenotype. Seefelder _et al_.25 were the first to construct a genetic linkage map of the male
hop, and they found 24 molecular markers which were sex specific. Danilova and Karlov26 were successful in the development of male specific molecular markers with the inter-simple sequence
repeat polymerase chain reaction (ISSR-PCR) method on hop plants of Russian and European origin. Čerenak _et al_.27 published a genetic linkage map where simple-sequence repeat (SSR) marker
HlAGA7 mapped to the male locus. Jakše _et al_.28 further described the HlAGA7 marker which appeared to distinguish male, female and monoecious plants in two Slovenian populations. In the
study of McAdam _et al_.29 the marker HlAGA7 showed complete linkage to the male sex phenotype in a New Zealand mapping population. Even though the marker proved to be perfectly associated
with the male sex, the use of this particular marker was limited due to the technical difficulties of using SSR with multiple alleles. Buck _et al_.30 developed four sex linked markers;
three sequenced characterised amplified region (SCAR) markers, and one high-resolution melting (HRM) curve analysis marker the latter of which has been successfully applied within the New
Zealand hop breeding programme31. Recently, Hill _et al_.22 identified a pseudoautosomal region (PAR), and male-specific regions of the Y-chromosome, along with genes located in these parts
of the sex chromosomes. In order to develop an efficient method to determine the sex phenotype among young seedlings in a hop breeding program, we developed a multiplex PCR approach
amplifying four male specific markers and a chloroplast specific DNA fragment. Male specific markers were idenitified from a DArT (Diversity Arrays Technology32 genetic mapping study33.
Selected DArT markers and the developed multiplex PCR approach were validated across a broad spectrum of hop genetic resources, in progeny of specific crosses, and for routine application in
the Slovene breeding program. Furthermore, we evaluated these markers on crude sap samples to avoid the tedious step of DNA isolation, with promising results. MATERIALS AND METHODS
PHENOTYPIC DETERMINATION OF SEX IN HOP PLANTS Sex phenotype was determined by in-field visual observation of seedlings during the flowering phase (from June to July) in years 2014 and 2015,
on 1 and 2 year old plants (respectively). The results of phenotypic sex determination were compared against the molecular markers. DNA EXTRACTION Total genomic DNA was extracted from fresh
plant material (leaves, plant buds, and tissue cultures) according to the modified CTAB protocol34. DNA concentration was quantified by means of fluorometry and samples stored at −20 °C.
MALE SPECIFIC DART MARKER DISCOVERY PLANT MATERIAL Three mapping populations from: (i) New Zealand (‘Nugget’ × ‘SBL3/3’, 170 plants30); (ii) Slovenia (‘Hallertauer Magnum’ × ‘SBL2/1’, 92
plants27,29); and (iii) USA (‘Perle’ × ‘USDA19058M’, 124 plants35) were included in sex linked marker development. DART MALE LINKED MARKERS In the framework of the hop DArT consortium
mapping project, all markers were sequenced by Sanger technology. The genotyping data for the Slovenian population were searched for DArT markers showing complete linkage with male sex
phenotype, while being absent in female plants (i.e., no recombination between the DArT marker and trait of interest). The resulting set of markers was compared against the phenotypic data
from the other two mapping populations, to infer male plants and to identify any additional male sex-linked markers. Identified male linked sequences were edited and assembled in CodonCode
Aligner (ver. 7.1.2) and submitted to GenBank (MG744425-MG744432). A hop chloroplast specific sequence named contig18 (GenBank MG744433) was obtained from our recent hop transcriptome
project36. PRIMER CONSTRUCTION Primer pairs for use in a single PCR were constructed using PRIMER337 web version using the default program parameters. Primers for the multiplex approach were
designed using the MPprimer tool38 specially developed to account for multiplex conditions, using default program options. Developed primers are presented in Tables 1 and 2. SINGLE PCR
MARKER REACTION PLANT MATERIAL 203 different genotypes (including cultivars, wild female and male plants, and monoecious plants; Supplementary Information 1) and 100 breeding lines (Table 3)
with known sex phenotypes from three different families were used in PCR amplification. PCR AMPLIFICATION The amplification was performed in 20 μl solution containing 1x PCR buffer, 2 mM
MgCl2, 0.2 mM each dNTPs, 0.5 μM primers, 0.5 U of _Taq_ DNA polymerase and 20 ng of genomic hop DNA using the following thermal cycling protocols: 1) primers hPb-CONT: 94 °C for 5 min,
followed by 40 cycles of 30 sec at 94 °C, 30 sec at 58 °C and 90 sec at 72 °C, 2) primers hPb-719005 (touchdown protocol): 94 °C for 5 min, followed by 10 cycles of 30 sec at 94 °C, 30 sec
at 62 °C (decreased -1 °C each cycle), 1 min at 72 °C and 30 cycles at 94 °C for 30 sec, 52 °C for 30 sec and 72 °C for 1 min, 3) primers hPb-365890 and hPb-718821: 94 °C for 5 min, followed
by 40 cycles of 30 sec at 94 °C, 30 sec at 67 °C and 1 min at 72 °C. All reactions were completed by incubating at 72 °C for 8 min. Only two primers, hPb-CONT and hPb-719005 (Table 1) were
further used for amplification across three crossing families (Table 3). Amplified PCR products were separated on 2% agarose gel and visualized by ethidium bromide staining. MULTIPLEX PCR
MARKER DEVELOPMENT PLANT MATERIAL A total of 97 hop accessions of different origins (Supplementary Information 1) were included in the optimization of multiplex PCR reaction. The sample set
comprised of 24 male and 73 female genotypes. Additional samples from three families (Table 3) were used for comparison of the results obtained from single and multiplex PCR reactions.
MULTIPLEX PCR AMPLIFICATION Single primer-pair PCRs were carried out initially, for each of the five multiplex primer pairs listed in Table 2. After initial confirmation by single pair
amplification, the multiplex amplification was optimized by varying the primer concentration. Optimized PCR conditions in 15 μl reactions were as follows: 40 ng DNA, 1x QIAGEN Multiplex PCR
Master Mix, 1x Q-Solution and primers at following concentrations: 0.2 μM for primers hPb-CONT, hPb-365890 and hPb-719005, 0.4 μM for primer hPb-718821 and 0.04 μM for primer contig18.
Different primer concentrations are crucial to achieving a comparable rate of amplification of five different fragments. For example, contig18 which is of chloroplast origin requires much
lower primer concentration, due to higher number of copies present in nucleic acid extract compared to the nuclear DNA. Amplification was carried out using the following thermal cycling
touchdown protocol: 95 °C for 15 min, followed by 8 cycles of 30 sec at 94 °C, 90 sec at 65 °C (decreased 1 °C each subsequent cycle) and 90 sec at 72 °C. The amplification continued for 27
cycles at 94 °C for 30 sec, 57 °C for 90 sec and 72 °C for 90 sec. The reactions were completed by incubation at 72 °C for 10 min. PCR products were separated on 2% agarose gel and
visualized by ethidium bromide staining. CRUDE SAMPLE MULTIPLEX PCR AMPLIFICATION PLANT MATERIAL As a first optimisation step, 10 samples of female varieties and 10 male plants were used for
amplification of crude sample PCR. Afterwards, 253 hop plants at the seedling stage from Slovenian hop breeding program representing 14 crossing families were analysed by using crude sample
multiplex PCR amplification (Table 4). CRUDE SAMPLE MULTIPLEX PCR AMPLIFICATION Crude sample multiplex PCR amplification was developed using Kapa 3 G Plant PCR kit (Kapa Biosystems)
utilizing fast extraction of crude DNA extract. Leaf disc circles (1 cm diameter) were excised by a puncture tool and immersed in 200 μl of extraction buffer (0.5 M Tris-HCl, 1 mM EDTA (pH =
8.0), 2% β-mercaptoethanol) with two steel beads (5 mm). Tissue was homogenized in TissueLyser (Qiagen) with 10 rotations per second for 30 sec. Samples were heated at 95 °C for 5 min,
cooled (-20 °C) for 10 min and centrifuged at 12,000 g for 10 min. Supernatant of crude extract was diluted in ratio 1:9 in sterile dH2O. Two μl of diluted crude DNA extract was used in 10
μl PCR solution containing 1x PCR buffer, 1.25 mM MgCl2, Enhancer (diluted 1:50), 0.2 U of KAPA3G Plant DNA polymerase and primers in same concentration as determined previously. The PCR
amplification profiles and agarose gel electrophoresis analysis were the same as described for multiplex reactions (see Multiplex PCR amplification). RESULTS DART MALE LINKED MARKERS
DISCOVERY In the Slovenian mapping family (‘Hallertauer Magnum’ × ‘SBL2/1’) represented by 92 plants, 9 were phenotypically male. Based on this observation, 10 DArT markers (hPb-361327,
hPb-363461, hPb-365890, hPb-366371, hPb-715987, hPb-716314, hPb-718821, hPb-718886, hPb-719005, and hPb-716926) were discovered that were present in the male parent, absent in the female
parent and present in all male siblings. Based on this information these markers were searched in the New Zealand (‘Nugget’ × ‘SBL3/3’) and USA (‘Perle’ × ‘USDA19058M’) mapping populations
for being present in male parent and in male siblings. Three markers (hPb-365890, hPb-716314 and hPb-719005) were confirmed to be present in all three families, 5 were common between
Slovenian and New Zealand’s families (hPb-718886, hPb-718821, hPb-715987, hPb-363461, and hPb-361327), while one marker was unique to each of the Slovenian family (hPb-716926) and USA family
(hPb-366371). Therefore, there were a total of 10 DArT markers specific to the male phenotype among those three families (Table 5). For two DArT markers, hPb-715987 and hPb-716314, quality
DNA sequences were not obtained, and therefore they were omitted from analysis. Comparison of the remaining eight male linked DArT marker sequences revealed that three markers hPb-361327,
hPb-363461 and hPb-718886, are almost identical showing three A- > G transitions and probably representing two alleles (Supplementary Information 2: Alignment of sequences). The primer
pair developed based on their alignment was named hPb-CONT; the other primers retained the DArT marker nomenclature. Together, six unique, male linked sequences were further tested in single
PCR amplification. INITIAL SINGLE PCR SCREENING OF MALE LINKED MARKERS Single PCR primers were developed (Table 1) and PCR conditions optimized on a set of four female and four male hops
comprising cultivars ‘Magnum’, ‘Perle’, ‘Comet’, and ‘Fuggle’ and male breeding lines 2/1, 3/3, 19058, and 29-70-54 for four DArT markers, including the contiguous sequence hPb-CONT
(representing hPb-361327, hPb-363461, hPb-718886), hPb-365890, hPb-718821 and hPb-719005. These four male-specific markers were further screened in 122 female genotypes (117 cultivars and 5
wild hops), 44 male genotypes and 37 monoecious genotypes (20 predominantly male phenotype – Mf; 16 predominantly female phenotype – Fm and 1 plant in which neither male nor female flowers
clearly predominate - FM - Supplementary Information 1). In summary, no male specific marker was successfully amplified in female genotypes, while the success of identifying males varied.
For example, markers hPb-CONT and hPb-365890 each failed to amplify in two male genotypes (hPb-CONT: No3-38 and 284/113; hPb-365890: 19058 and 120/13). The marker hPb-718821 was not
amplified in one male genotype (19058), and hPb-719005 did not amplify in four males (No3-38, 85/169, 19058 and 120/13). Interestingly, male specific markers were amplified in Mf
hermaphroditic plants while not in Fm plants. For two DArT marker sequences (hPb-366371 and hPb-716926) amplification could not be achieved without varying the PCR conditions. Further
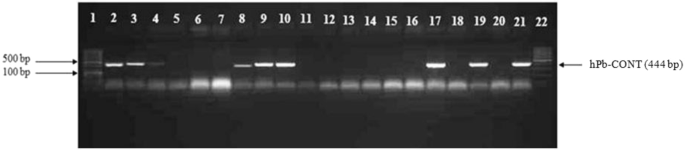
analysis was performed on 100 breeding lines from 3 crossing families (Table 3) by using primers hPb-719005 and hPb-CONT (Fig. 1). By using primers hPb-719005 and hPb-CONT the correct sex
determination was achieved between 94% and 100% of cases, depending upon the marker used and the family, showing the importance of using all four male specific markers. Therefore, all 5
markers are required to maximise the likelihood that the assay is effective throughout the biogeographic range of hop. This is important as the large majority of hop in commercial production
and breeding programs around the world is some form of multi-generation hybrid between European and North American germplasm. DEVELOPMENT OF MULTIPLEX PCR APPROACH Since neither hPb-CONT
nor hPb-719005 were able to detect 100% known males independently, we aimed to develop a multiplex PCR approach where all four markers could be amplified simultaneously. In multiplex primer
development the fifth sequence of hop chloroplast origin (contig18) was included to confirm amplification of isolated DNA and to exclude the risk of false negatives being identified as
females. The developed multiplex primer set is presented in Table 2. During multiplex optimization the duplex PCR amplifications were performed by using non-sex specific chloroplast primer
contig18 and four primers linked to male sex phenotype (hPb-CONT, hPb-719005, hPb-365890, hPb-718821 - Fig. 2). The verification of multiplex PCR amplification was initially checked on 97
genotypes (73 female and 24 male plants) representatives of globally significant hop accessions (Supplementary Information 1) by multiplying all five primers (hPb-CONT, hPb-365890,
hPb-719005, hPb-718821, contig18). The markers successfully predicted sex phenotype in 97/97 cases (Fig. 3). In one male plant, a wild male genotype (63012 - Fig. 3, lane 20) of North
American biogeographic origin, the primer hPb-718821 failed to amplify the fragment, even when the reaction was repeated. Nevertheless, the sex phenotype of the plant was correctly
distinguished based on amplification of the other three male specific markers. The results obtained from single PCR amplification and multiplex PCR amplification were compared. By using
hPb-CONT and hPb-719005 primers, the results of sex determination in three crossing families coincided with phenotypic determination from 94% to 96% and 94% to 100%, respectively.
Comparatively, when multiplex of five primers for PCR amplification was used in same progeny, 100% success was achieved. Furthermore, correct results were obtained by using multiplex PCR in
the analyses of 96 world hop accessions (Supplementary Information 1). DIRECT PCR REACTION USING CRUDE NUCLEIC ACID EXTRACT With the aim of accelerating sex determination in the hop breeding
process, the possibility of using crude sample multiplex PCR amplifications was investigated on an initial 20 samples. In this initial test, the multiplexed markers correctly diagnosed the
sex phenotype of each sample (Fig. 4). Furthermore, in 14 families (a total of 253 genotypes – Table 4) the sex phenotype was diagnosed by crude sample multiplex PCR at seedling stage. After
molecular analysis the plants were separately planted according to their marker determined sex genotype, in field trials within the context of the Slovenian hop breeding program. In the
subsequent 1–2 years, phenotypic observations were collected and the comparison with molecular data was obtained. As can be seen from Table 4, the results from crude multiplex PCR
amplification coincided perfectly with the in-field determination of plant sex phenotype. DISCUSSION In dioecious species, where plants of one sex phenotype are preferred for commercial use,
breeding or cultivation, various DNA fingerprinting methods have been employed for sex determination, as described in species such as _Cannabis sativa_39, _Asparagus officinalis_40,41,
_Actinidia chinensis_42, and _Ficus fulva_43. In some species such as _Garcinia gummi-gutta_44, _Simmondsia chinensis_45, and _Calamus guruba_11, the time between germination and the onset
of reproductive maturity may be several years, imposing a strong limitation on the use of phenotypic determination of sex9. In hop, male plants cannot be differentiated phenotypically from
female plants before flowering (sexual maturity), early sex determination via molecular markers is appealing. Beatson _et al_.31 reported the economic benefits of using a HRM sex marker for
elimination of male seedlings at the nursery stage in several New Zealand triploid breeding populations. The sex determination system is similar in _Humulus, Cannabis_ and _Rumex_ species,
where the XX/XY system exists and the ratio between X chromosomes and autosomes effects sex expression8,19. In the family Cannabaceae, the three species – _Cannabis sativa, Humulus lupulus_
and _Humulus japonicus_ – are dioecious with heterogametic sex chromosomes. In _H. lupulus_, Ono46 described six different systems, where 1 to 3 pairs of sex chromosomes with different sizes
of Y chromosome exist. Divashuk _et al_.47 reported a cytogenetic marker for the identification of sex chromosomes in _H. lupulus_. They revealed pseudo autosomal regions on the long arms
of the X and Y chromosomes. Furthermore, Hill _et al_.22 identified a set of loci that are sex-linked and probably located in the pseudo autosomal region. Their study identified a 1.3 Mb
section of DNA that appears unique to male hop genotypes. They proposed that the identified region could be utilised for the development of molecular markers for diagnosis of sex phenotype.
All reported sex determining molecular markers in hop, have been linked to the maleness. Polley _et al_.6 were the first to publish a sex-specific DNA sequence in hop, developed from random
amplified polymorphic DNA (RAPD) molecular markers by using bulk segregant analysis (BSA), which is predominantly present on the Y chromosome and hybridized only weakly to female DNA. Upon
testing5,23, this first sex-linked marker did not appear to coincide completely with the phenotypic assessments. Jakše _et al_.28 reported an SSR marker tightly linked to the male sex in
hop, which appeared to show complete linkage to the male character but technical difficulties associated with SSR genotyping meant that this marker was not applied in marker assisted
selection (MAS). Hop breeding programs are long lasting, with a development timeline of 10 to 15 years from crossing to registration of a new cultivar. To improve the efficiency of
traditional selection procedures, several different molecular markers have been used to analyse genetic distances among hop breeding genotypes33,35,48,49,50, and to detect QTLs (quantitative
trait loci)26,27,28,51,52. Nevertheless, implementation of developed techniques in marker-assisted selection has remained unpublished up to this point. In order to avoid the delayed
identification of male plants within the experimental systems of hop genetic improvement, we developed and applied selected DArT molecular markers to determine sex at an early hop seedling
stage. The present study was based on the research of Howard _et al_.33, where 730 polymorphic markers from 92 hop accessions were discovered using diversity arrays technology (DArT), which
were further used in linkage studies29. There have been few studies describing the transferability of DArT markers identified through research, into breeding programs. In most cases markers
were used in variability studies and genome mapping in different species, such as olive (_Olea europaea_ L.)53,54, sugar beet (_Beta vulgaris_ L.)55, apple (_Malus domestica_ Borkh.)56,
eucalypt (_Eucaplyptus_ spp.)57,58, hexaploid wheat (_Triticum aestivum_ L.)59,60, perennial ryegrass (_Lolium perenne_ L.)61, and tomato (_Solanum lycopersicum_ L.)62. By studying sex
linked marker performance in 197 plants (97 from the global hop genotype collection and 100 from crossing families), sex determined by multiplex PCR amplification appears to completely
coincide with in-field phenotypic assessment. Furthermore, by using a crude sample multiplex PCR technique, the determination of sex in hop seedlings was made more efficient, without losing
accuracy. The present research verified both single, and multiplex PCR sex-linked markers in 203 genotypes from world collection and three experimental families and developed and
demonstrated the efficacy of a system of rapid, crude sample extract multiplex PCR sex-linked markers among progeny of 14 crosses. To conclude, selected markers combined into a crude sample
multiplex PCR assay, were tested in a broad spectrum of hop genetic resources, in progeny of crosses, and for routine application around 4.000 seedlings were annualy tested in the period of
2015–2018 in an active hop breeding program. It is important to note that taking into consideration all technical hours previously occupied in preventing male plants from flowering near
commercial fields and the fact that accurate determination of sex at the seedling stage reduces the trial area required for screening by about one third, the financial cost of MAS has been
recovered. Further, the laboratory analysis can be performed during winter, using leaf samples collected the previous growing season, spreading research activity away from seasonal labour
peaks associated with hop production. The methods described above appear to produce complete linkage between multiplex PCR sex-linked molecular markers and phenotypic sex expression in field
grown hop, and would appear to be appropriate for routine testing of hop seedlings in breeding programs worldwide. REFERENCES * Zhao, F. _et al_. Prenylflavonoids and phloroglucinol
derivatives from hops (_Humulus lupulus_). _J Natur Prod_ 68, 43–49 (2005). Article CAS Google Scholar * Mikyška, A., Hrabák, M., Hašková, D. & Šrogl, J. The role of malt and hop
polyphenols in beer quality, flavour and haze stability. _J Inst Brew_ 108(1), 78–85 (2002). Article Google Scholar * Milligan, S. _et al_. Oestrogenic activity of the hop phyto-oestrogen,
8-prenylnaringenin. _Reproduction_ 123(2), 235–242 (2002). Article CAS Google Scholar * IHGC. IHGC (International Hop Growers’ Convention)
http://www.hmelj-giz.si/ihgc/doc/2018%20MAY%20IHGC%20EC%20Reports.pdf (29.8.2018) (2018). * Patzak, J., Vejl, P., Skupinová, S. & Nesvadba, V. Identification of sex in F progenies of hop
(_Humulus lupulus_ L.) by molecular marker. _Rost Vyroba_ 48(7), 318–321 (1999). Google Scholar * Polley, A., Ganal, M. & Seigner, E. Identification of sex in hop (_Humulus lupulus_)
using molecular markers. _Genome_ 40(3), 357–361 (1997). Article CAS Google Scholar * Thomas, G. G. & Neve, R. A. Studies on the effect of pollination on the yield and resin content
of hops (_Humulus lupulus_). _J Inst Brew_ 82, 41–45 (1976). Article CAS Google Scholar * Juarez, C. & Ann Banks, J. Sex determination in plants. _Curr Opin Plant Biol_ 1(1), 68–72
(1998). Article CAS Google Scholar * Kumar, S., Kumari, R. & Sharma, V. Genetics of dioecy and causal sex chromosomes in plants. _J Genet_ 93(1), 241–277 (2014). Article Google
Scholar * Heikrujam, M., Sharma, K., Kumar, J. & Agrawal, V. Generation and validation of unique male sex-specific sequence tagged sites (STS) marker from diverse genotypes of dioecious
_Jojoba-Simmondsia chinensis_ (Link) Schneider. _Euphytica_ 199, 363–372 (2014). Article Google Scholar * Renner, S. S. & Ricklefs, R. E. Dioecy and its correlates in the flowering
plants. _Am J Bot_ 82, 596–606 (1995). Article Google Scholar * Sinha, P., Nanda, S., Joshi, R. K. & Panda, P. C. Development of a sequence-tagged site (STS) marker for sex
identification in the dioecious rattan species Calamus guruba Buch. -Ham. Mol Breed 37(22), 10.1007/s11032-017-0630-z (2017). * Mohanty, J., Nayak, S., Jha, S. & Joshi, R. A sequence
tagged site (STS) marker encoding Copia-like retrotransposable element is associated with male specific sex expression in _Momordica dioica Roxb_. _Sci Hortic_ 201, 265–270 (2016). Article
CAS Google Scholar * Bhowmick, B., Nanda, S., Nayak, S., Jha, S. & Joshi, R. An APETALA3 MADS-box linked SCAR marker associated with male specific sex expression in _Coccinia grandis_
(L). _Voigt Sci Hortic_ 176, 85–90 (2014). Article CAS Google Scholar * Gangopadhyay, G. _et al_. Sex detection of _Carica papaya_ and _Cycas circinalis_ in pre-flowering stage by ISSR
and RAPD. _Curr Sci_ 92(4), 524–526 (2007). CAS Google Scholar * Törjék, O. _et al_. Novel male-specific molecular markers (MADC5, MADC6) in hemp. _Euphytica_ 127, 209–218 (2002). Article
Google Scholar * Neve, R. A. Hops. Chapman and Hall, London (1991) * Shepard, H., Parker, J., Darby, P. & Ainsworth, C. Sexual development and sex chromosomes in hop. _New Phytol_
148(3), 397–411 (2000). Article Google Scholar * Karlov, G. I., Danilova, T. V., Horlemann, C. & Weber, G. Molecular cytogenetics in hop (_Humulus lupulus_ L.) and identification of
sex chromosomes by DAPIbanding. _Euphytica_ 132, 185–190 (2003). Article CAS Google Scholar * Parker, J. & Clark, M. Dosage sex-chromosome systems in plants. _Plant Sci_ 80, 79–92
(1991). Article Google Scholar * Dellaporta, S. L. & Calderon-Urrea, A. Sex determination in flowering plants. _Plant Cell_ 5, 1241–1251 (1993). CAS PubMed PubMed Central Google
Scholar * Hill, S. T., Coggins, J., Liston, A., Hendrix, D. & Henning, J. A. Genomics of the hop pseudo-autosomal regions. _Euphytica_ 209, 171–179 (2016). Article CAS Google Scholar
* Škof, S., Čerenak, A., Jakše, J., Bohanec, B. & Javornik, B. Ploidy and sex expression in monoecious hop (_Humulus lupulus_). _Botany_ 90(7), 617–626 (2012). Article Google Scholar
* Čerenak, A. & Javornik, B. Application of male STS marker in hop (Humulus lupulus L.) breeding. V: Seigner E… (ed). _Proceedings of the Scientific Commission [of the] International
Hop Growers’ Convention I.H.G.C_. [S. l.]: International Hop Growers’ Convention 39–42 (1999). * Seefelder, S., Ehrmaier, H., Schweizer, G. & Seigner, E. Male and female genetic linkage
map of hops, _Humulus lupulus_. _Plant Breed_ 119(3), 249–255 (2000). Article CAS Google Scholar * Danilova, T. & Karlov, G. Application of inter simple sequence repeat (ISSR)
polymorphism for detection of sex-specific molecular markers in hop (_Humulus lupulus_ L.). _Euphytica_ 151(1), 15–21 (2006). Article CAS Google Scholar * Čerenak, A., Šatović, Z. &
Javornik, B. Genetic mapping of hop (_Humulus lupulus_ L.) applied to the detection of QTLs for alpha-acid content. _Genome_ 49, 485–494 (2006). Article Google Scholar * Jakše, J.,
Štajner, N., Kozjak, P., Čerenak, A. & Javornik, B. Trinucleotide microsatellite repeat is tightly linked to male sex in hop (_Humulus lupulus_ L.). _Mol Breed_ 21(2), 139–148 (2008).
Article Google Scholar * McAdam, E. L. _et al_. Quantitative trait loci in hop (_Humulus lupulus_ L.) reveal complex genetic architecture underlying variation in sex, yield and cone
chemistry. _BMC Genomics_ 14(1), 1 (2013). Article Google Scholar * Buck, E. J., _et al_ The development and mapping of four new genetic markers for gender determination in hop (_Humulus
lupulus_ L). _Plant & Animal Genomes XVIII Conference_, 9–13 January 2010, San Diego, California (2010). * Beatson, R. A. _et al_. Breeding polyploid hop cultivars for New Zealand
conditions. _Acta Hortic_ 1127, 9–14 (2016). Article Google Scholar * Jaccoud, D., Peng, K., Feinstein, D. & Kilian, A. Diversity arrays: a solid state technology for sequence
information independent genotyping. _Nucleic Acids Res_ 2001(29), E25 (2001). Article Google Scholar * Howard, E. _et al_. High-throughput genotyping of hop (_Humulus lupulus_ L.)
utilising diversity arrays technology (DArT). _Theor Appl Genet_ 122(7), 1265–1280 (2011). Article CAS Google Scholar * Kump, B. & Javornik, B. Evaluation of genetic variability among
common buckwheat (_Fagopyrum esculentum_ Moench.) populations by RAPD markers. _Plant Sci_ 114, 149–158 (1996). Article CAS Google Scholar * Henning, J. A. _et al_. QTL mapping of
powdery mildew susceptibility in hop (_Humulus lupulus_ L.). _Euphytica_ 180(3), 411–420 (2011). Article MathSciNet Google Scholar * Pokorn, T., Radišek, S., Javornik, B., Štajner, N.
& Jakše, J. Development of hop transcriptome to support research into host-viroid interactions. _PLoS One_ 12(9), e0184528, https://doi.org/10.1371/journal.pone.0184528 (2017). Article
CAS PubMed PubMed Central Google Scholar * Rozen, S. & Skaletsky, H. Primer3 on the WWW for General Users and for Biologist Programmers. In: Bioinformatics Methods and Protocols, vol
132. _Methods in Mol Biol_. 365–386 (1999). * Shen, Z. _et al_. MPprimer: a program for reliable multiplex PCR primer design. _BMC Bioinformatics_ 11(1), 143,
https://doi.org/10.1186/1471-2105-11-143 (2010). Article CAS PubMed PubMed Central Google Scholar * Flachowsky, H., Schumann, E., Webbere, W. E. & Peil, A. Application of AFLP for
the detection of sex-specific markers in hemp. _Plant Breed_ 120, 305–309 (2001). Article CAS Google Scholar * Jamsari, A., Nitz, I., Reamon-Buttner, S. M. & Jung, C. BAC-derived
diagnostic markers for sex determination in asparagus. _Theor Appl Genet_ 108, 1140–1146 (2004). Article CAS Google Scholar * Telgmann-Rauber, A., Jamsari, A., Kinney, M. S., Pires, J. C.
& Jung, C. Genetic and physical maps around the sex-determining M-locus of the dioecious plant asparagus. _Mol Genet Genomics_ 278, 221–234 (2007). Article CAS Google Scholar *
Shirkot, P., Sharma, D. & Mohapatra, T. Molecular identification of sex in _Actinidia deliciosa_ var. _deliciosa_ by RAPD markers. _Sci Hortic_ 94(1–2), 33–39 (2002). Article CAS
Google Scholar * Parrish, T. L., Koelewijn, H. P. & van Dijk, P. J. Identification of a male-specific AFLP marker in a functionally dioecious fig, _Ficus fulva_ Reinw. ex Bl.
(Moraceae). _Sex Plant Reprod_ 17, 17–22 (2004). Article CAS Google Scholar * Joseph, K. S., Murthy, H. N. & Ravishankar, K. V. Development of SCAR marker for sex identification in
dioecious _Garcinia gummi-gutta_. _Trees_ 28, 1645–1651 (2014). Article CAS Google Scholar * Sharma, K., Agrawal, V., Gupta, S., Kumar, R. & Prasad, M. ISSR marker-assisted selection
of male and female plants in a promising dioecious crop: jojoba (_Simmondsia chinensis_). _Plant Biotechnol Rep_ 2, 239–243 (2008). Article Google Scholar * Ono, T. Chromosomes of common
hop and its relatives. _Bull Brew Sci_ 2, 3–65 (1961). Google Scholar * Divashuk, M. G., Alexandrov, O. S., Yu Kroupin, P. & Karlov, G. I. Molecular Cytogenetic mapping of _Humulus
lulupus_ sex chromosomes. _Cytogenet Genome Res_ 134, 213–219 (2011). Article CAS Google Scholar * Hartl, L. & Seefelder, S. Diversity of selected hop cultivars detected by
fluorescent AFLPs. _Theor Appl Genet_ 96, 112–116 (1998). Article CAS Google Scholar * Henning, J. H., Townsend, M. S. & Matthews, P. Predicting offspring performance in hop (_Humulus
lupulus_ L.) using AFLP markers. _J Am Soc Brew Chem_ 68, 125–131 (2010). CAS Google Scholar * Matthews, P. D., Coles, M. C. & Pitra, N. J. Next generation sequencing for a plant of
great tradition: Application of NGS to SNP detection and validation in hops (_Humulus lupulus_ L.). _Brew Sci_ 66, 185–191 (2013). Google Scholar * Čerenak, A. _et al_. Identification of
QTLs for alpha acid content and yield in hop (Humulus lupulus L.). _Euphytica_ 170(1–2), 141–154 (2009). Article Google Scholar * Koie K, Inaba A, Okada Y, Kaneko T, Ito K. Construction of
the genetic linkage map and QTL analysis on hop (_Humulus lupulus_ L.). In _Proceedings of the 1_ _st_ _International Humulus Symposium held_ 1–7 Aug. 2004, Corvallis, Oregon. _Edited by_
K. E. Hummer and J. A. Henning. _Acta Hort_ 668, ISHS 2005:59–67 (2005). * Atienza, S. G. _et al_. Use of DArT markers as a means of better management of the diversity of olive cultivars.
_Food Res Int_ 54, 2045–2053 (2013). Article CAS Google Scholar * Domínguez-García, M. C. _et al_. Development of DArT markers in olive (_Olea europaea_ L.) and usefulness in variability
studies and genome mapping. _Sci Hortic_ 136, 50–60 (2012). Article Google Scholar * Simko, I., Eujayl, I. & van Hintum, T. J. Empirical evaluation of DArT, SNP, and SSR marker-systems
for genotyping, clustering, and assigning sugar beet hybrid varieties into populations. _Plant Sci_ 184, 54–62 (2012). Article CAS Google Scholar * Schouten, H. J. _et al_. Diversity
arrays technology (DArT) markers in apple for genetic linkage maps. _Mol Breed_ 29, 645–660 (2012). Article CAS Google Scholar * Kullan, A. R. K. _et al_. High-density genetic linkage
maps with over 2,400 sequence-anchored DArT markers for genetic dissection in an F pseudo-backcross of _Eucalyptus grandis_ × _E. urophylla_. _Tree Genet Genomes_ 8, 163–175 (2011). Article
Google Scholar * Larcombe, M. J. _et al_. Patterns of reproductive isolation in _Eucalyptus_ – a phylogenetic perspective. _Mol Biol Evol_ 32(7), 1833–1846 (2015). Article CAS Google
Scholar * Akbari, M. _et al_. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. _Theor Appl Genet_ 113(8), 1409–1420 (2006). Article CAS
Google Scholar * Francki, M. G. _et al_. Comparison of genetic and cytogenetic maps of hexaploid wheat (_Triticum aestivum_ L.) using SSR and DArT markers. _Mol Genet Genomics_ 281, 181–191
(2009). Article CAS Google Scholar * King, J., Thomas, A., James, C., King, I. & Armstead, I. A DArT marker genetic map of perennial ryegrass (_Lolium perenne_ L.) integrated with
detailed comparative mapping information; comparison with existing DArT marker genetic maps of _Lolium perenne_, _L. multiflorum_ and _Festuca pratensis_. _BMC Genomics_ 14(1), 437 (2013).
Article CAS Google Scholar * Van Schalkwyk, A. _et al_. Bin mapping of tomato diversity array (DArT) markers to genomic regions of _Solanum lycopersicum_ x _Solanum pennellii_
introgression lines. _Theor Appl Genet_ 124, 947–956 (2012). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS DArT marker development was supported by Horticulture
Australia projects OT04003 and HP08002. The authors acknowledge financial support from the Slovenian Research Agency P4-0077, 6316-3/2011-784 (Z.K.) and 1000-09-212225 (S.Š.). The Slovenian
hop breeding program is supported by the Slovenian Ministry of Agriculture, Forestry and Food and Slovenian hop farmers. New Zealand populations were contributed by The New Zealand Institute
for Plant & Food Research Limited and supported by a New Zealand Ministry for Business Innovation & Employment research contract C11X1006. AUTHOR INFORMATION AUTHORS AND
AFFILIATIONS * Slovenian Institute of Hop Research and Brewing, Cesta Žalskega tabora 2, 3310, Žalec, Slovenia Andreja Čerenak, Zala Kolenc & Petra Sehur * Hop Products Australia, 446
Elizabeth Street Hobart, Tasmania & School of Natural Sciences, University of Tasmania, Private Bag 55, Hobart, Tasmania, Australia Simon P. Whittock * University of Tasmania, School of
Natural Sciences, Private Bag 55, Hobart, TAS, 7001, Australia Anthony Koutoulis * The New Zealand Institute for Plant & Food Research Limited, Palmerston North Research Centre, Private
Bag 11600, Palmerston North, 4442, New Zealand Ron Beatson & Emily Buck * University of Ljubljana, Biotechnical Faculty, Agronomy Department, Jamnikarjeva 101, 1000, Ljubljana, Slovenia
Branka Javornik, Suzana Škof & Jernej Jakše Authors * Andreja Čerenak View author publications You can also search for this author inPubMed Google Scholar * Zala Kolenc View author
publications You can also search for this author inPubMed Google Scholar * Petra Sehur View author publications You can also search for this author inPubMed Google Scholar * Simon P.
Whittock View author publications You can also search for this author inPubMed Google Scholar * Anthony Koutoulis View author publications You can also search for this author inPubMed Google
Scholar * Ron Beatson View author publications You can also search for this author inPubMed Google Scholar * Emily Buck View author publications You can also search for this author inPubMed
Google Scholar * Branka Javornik View author publications You can also search for this author inPubMed Google Scholar * Suzana Škof View author publications You can also search for this
author inPubMed Google Scholar * Jernej Jakše View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS J.J., A.Č., S.P.W., R.B., E.B., B.J. and A.K.
planned and designed the research. A.K., S.P.W., E.B. and J.J. analysed the DArT data. Z.K., P.S. and S.Š. conducted the laboratory work. A.K., S.P.W., A.Č. and J.J. interpreted the data
and drafted the manuscript. All authors confirmed the final version of the manuscript. CORRESPONDING AUTHOR Correspondence to Andreja Čerenak. ETHICS DECLARATIONS COMPETING INTERESTS The
authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License,
which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link
to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless
indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or
exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints
and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Čerenak, A., Kolenc, Z., Sehur, P. _et al._ New Male Specific Markers for Hop and Application in Breeding Program. _Sci Rep_ 9, 14223
(2019). https://doi.org/10.1038/s41598-019-50400-z Download citation * Received: 08 January 2019 * Accepted: 06 September 2019 * Published: 02 October 2019 * DOI:
https://doi.org/10.1038/s41598-019-50400-z SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative