Upf1 inhibits the hepatocellular carcinoma progression by targeting long non-coding rna uca1
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Hepatocellular carcinoma (HCC) is one of the major causes of cancer-related death worldwide. However, the molecular mechanism underlying HCC carcinogenesis remains to be further
elucidated. Up-frameshift protein 1 (UPF1) is a RNA/DNA-dependent ATPase and ATP-dependent RNA helicase. Here, we explored the expression and function of UPF1 in HCC. In this study, we
demonstrated that UPF1 expression was significantly reduced in hepatocellular carcinoma (HCC) tissues compared with the adjacent normal tissues. And further functional assays revealed that
knockdown of UPF1 promoted HCC cells growth and invasion. Furthermore, we found that UPF1 could bind to long non-coding RNA urothelial cancer associated 1 (UCA1) and was negatively
correlated with UCA1. UCA1 expression also affected HCC growth and invasion. Knockdown of UCA1 ameliorated the effect of UPF1 knock down on HCC growth and invasion. Knockdown of UPF1
enhances glycolysis in HCC. Taken together, our results provided new insights for finding novel therapeutic targets for hepatocellular carcinoma progression. SIMILAR CONTENT BEING VIEWED BY
OTHERS LNCRNA SNHG6 PROMOTES G1/S-PHASE TRANSITION IN HEPATOCELLULAR CARCINOMA BY IMPAIRING MIR-204-5P-MEDIATED INHIBITION OF E2F1 Article 06 April 2021 THE LIVER-SPECIFIC LONG NONCODING RNA
FAM99B INHIBITS RIBOSOME BIOGENESIS AND CANCER PROGRESSION THROUGH CLEAVAGE OF DEAD-BOX HELICASE 21 Article Open access 14 February 2025 LIN28B-AS1-IGF2BP1 BINDING PROMOTES HEPATOCELLULAR
CARCINOMA CELL PROGRESSION Article Open access 11 September 2020 INTRODUCTION Hepatocellular carcinoma (HCC) is one of the major causes of cancer-related death worldwide1. HCC results from
various types of diseases including liver fibrosis, liver cirrhosis, fatty liver and liver metastasis from other cancers2,3,4,5. There are so many risk factors leading to the malignant
transformation of the hepatocytes6,7,8. The etiology of HCC is so complicated that the details of the molecular mechanisms underlying HCC carcinogenesis remain to be further elucidated.
Up-frameshift protein 1 (UPF1) as a RNA/DNA-dependent ATPase and ATP-dependent RNA helicase is an evolutionarily conserved and ubiquitously expressed phosphoprotein9. UPF1 play a key player
in nonsense mediated mRNA decay (NMD) and non-NMD RNA degradation10. UPF1 associates with a translation termination codon by the translation termination complex during NMD. Moreover, UPF1
also promotes cell progression through G1/S raising the possibility that NMD promotes the decay of mRNAs encoding inhibitory proteins that block progression through this stage of the cell
cycle11. Recent study also found that the human RNA surveillance factor UPF1 regulates tumorigenesis12. In the present study, we demonstrated that UPF1 expression was significantly reduced
in hepatocellular carcinoma (HCC) tissues compared with the adjacent normal tissues. And further functional assays revealed that knockdown of UPF1 promoted HCC cells growth and invasion.
Furthermore, we found that UPF1 could bind to long non-coding RNA UCA1 and negatively correlated with UCA1. UCA1 expression also affected HCC growth and invasion. Knockdown of UCA1
ameliorated the effect of UPF1 knock down on HCC growth and invasion. Knockdown of UPF1 enhances glycolysis in HCC. Taken together, our results provided new insights for finding novel
therapeutic targets for hepatocellular carcinoma progression. MATERIALS AND METHODS CLINICAL SAMPLES Fifty pairs of liver cancer and adjacent non-tumor samples were collected from
hepatocellular cancer patients and stored in liquid nitrogen before use. The written informed consent was obtained from all patients before their inclusion in this study. The Medical Ethics
Committee of the Affiliated Hospital of Xi’an Medical University approved ethical approval for this study. All methods were performed in accordance with the relevant guidelines and
regulations of Medical Ethics Committee of the Affiliated Hospital of Xi’an Medical University. CELL CULTURE Human hepatocellular carcinoma Huh7 and HeG2 cells were cultured with RPMI 1640
medium supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco) in an atmosphere of 37 °C, 5% CO2.
IMMUNOHISTOCHEMISTRY (IHC) For IHC, the procedures were performed as the previous study13. Simply, the paraffin-embedded sections were deparaffinized, rehydrated. The sections were incubated
in anti-UPF1 antibody (Sigma-Aldrich, St. Louis, MO, USA) with a dilution of 1:200 overnight at 4 °C and then incubated with secondary antibody accordingly. Finally, a peroxidase substrate
was added and incubated until desired stain intensity developed. Tissue sections were mounted on slides using mounting medium. QUANTITATIVE REAL-TIME REVERSE TRANSCRIPTION-PCR Total RNA from
clinical samples was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and was reverse-transcribed using a PrimeScriptRT reagent Kit (Promega, Madison, WI, USA) according to
the manufacturer’s instruction. In brief, quantitative real-time reverse transcription -PCR was then performed using SYBR Green Master Mix (Thermo Fisher Scientific, Waltham, MA USA) and
ABI7900HT Real Time PCR system (Applied Biosystems, Foster City, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control. The primers used are as follows
GAPDH (5′-CCATGTTCGTCATGGGTGTGAACCA-3′ and 5′-GCCAGTAGAGGCAGGGATGATGTTG-3′), UPF1 (5′-ACCGACTTTACTCTTCCTAGCC-3′ and 5′-AGGTCCTTCGTGTAATAGGTGTC-3′) and UCA1 (5′- ATGCTTCCATGCAACACTCCT-3′ and
5′-GATTTTTTGTTTTGGGTGTGG-3′). The relative expression levels of each gene were calculated and determined using the 2−ΔΔct method. SIRNA SYNTHESIS AND CELL TRANSFECTION siRNAs targeting UPF1
or UCA1 was synthesized by the Ribobio (Guangzhou, China). The negative control siRNA was also purchased from Riobobio. Cells were transfected using Lipofectamine 2000 transfection reagent
(Thermo Fisher Scientific, Waltham, MA USA) according to the manufacturer’s instructions. At 48 hours after transfection, the cells were harvested, and the extracts were for further study.
Plasmids were also used to transfect as the same above method. METHYL-THIAZOLYL-TETRAZOLIUM (MTT) ASSAY Cells were plated onto 96-well plates before incubation overnight. The medium was
exchanged every 24 hours. The MTT (Sigma-Aldrich, St. Louis, MO, USA) was added to the cell supernatant, and removed after four hours at 37 °C. Then, 200 µl DMSO was added to each well and
shaken for 15 minutes. The optical density (OD) value was measured at 490 nm by an enzyme micro-plate reader. Each experiment was repeated three times. WESTERN BLOTTING Protein
concentrations of cell lysate were measured using a standard BCA assay according to the manufacturer’s instructions. Proteins were separated in SDS–PAGE (10%) and then transferred to
nitrocellulose membranes at 4 °C. Membranes were blocked with 5% nonfat dry milk in TBST for 1 hour, and then incubated overnight at 4 °C with the antibodies for UPF1 or GAPDH (Cell
Signaling, Danvers, MA). After washing with TBST, membranes were incubated with the secondary antibody for 1 hour at room temperature. The signals were developed with the ECL kit. COLONY
FORMATION Firstly, cells were plated in six-well plates (5 × 103 cells per well) before incubation overnight, and then cultured in RPMI 1640 medium for 2 weeks after transfection. After the
cells were washed with PBS twice, the cells were stained with 0.1% crystal violet. Finally the colonies with more than 50 cells were counted. TRANSWELL INVASION ASSAY The invasion assays
were performed by a Transwell chamber, according to the manufacturer’s protocol. Briefly, the membranes of each upper chamber of an insert were coated with Matrigel Basement Membrane Matrix
(BD Biosciences, Bedford, MA, USA) and then incubated for 5 h at 37 °C. Cells were seeded in the upper chamber with serum-free media, and the lower chamber was loaded with 700 μl of complete
medium. And then the cells were incubated for 24 h, washed, fixed, and then stained with crystal violet before counting under a microscope. RNA STABILITY ASSAY Huh7 and HepG2 cells
transfected with siRNA targeting for UPF1/UCA1 or control were incubated with 5 μg/ml of Actinomycin D (Sigma-Aldrich, St. Louis, MO, USA) in the medium. Total RNA was harvested at the
indicated time and then mRNA expression level was evaluated by real time PCR. The mRNA half-life was detected before and after adding Actinomycin D. RNA IMMUNOPRECIPITATION ASSAY RNA
immunoprecipitation (RIP) were carried out as described previously14. In brief, cells were harvested using a mixed buffer on ice for 20 min. Then nuclei were pelleted by centrifugation at
2,500 g for 15 min and then nuclear pellets were resuspended in RIP buffer. Nuclear membrane and debris were pelleted by centrifugation. Antibody to rabbit IgG or UPF1 (Cell Signaling,
Danvers, MA) was added to supernatant together with protein G beads (Thermo Fisher Scientific, Waltham, MA USA) and incubated overnight at 4 °C. Then co-precipitated RNAs were isolated and
qRT-PCR for UCA1 was conducted. GLUCOSE CONSUMPTION AND LACTATE PRODUCTION ASSAY To measure the levels of glucose and lactate, cell supernatants were collected and detected by a glucose and
lactate assay kit (BioVision, Milpitas, CA, USA) according to the manufacturer’s instructions. STATISTICAL ANALYSIS The statistical analyses were performed by SPSS 16 software. All the
experiments were performed three times, and the data were presented as the mean ± SD. The results were analyzed using one-way ANOVA and Student’s t-test. _P_ < 0.05 was considered
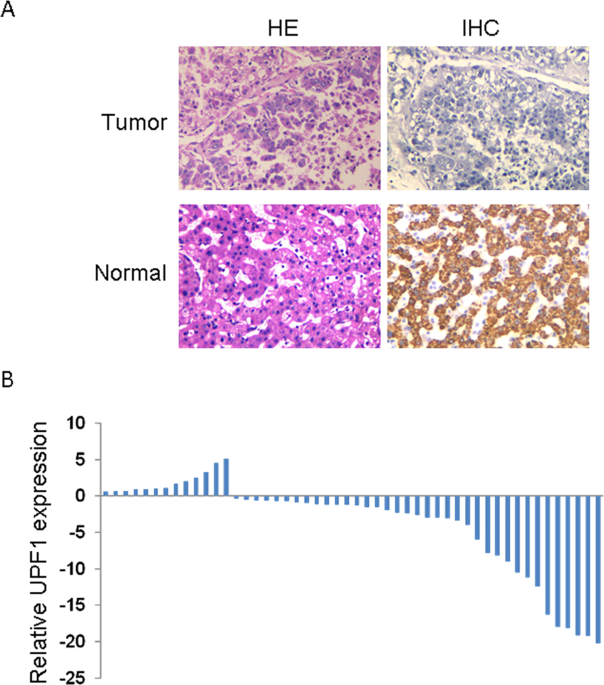
statistically significant. RESULTS UPF1 EXPRESSION WAS DECREASED IN HCC To investigate the expression and clinical significances of UPF1 in Hepatocellular Carcinoma (HCC), we first measured
UPF1 expression in 50 cancer tissues and paired adjacent noncancerous HCC tissues. As shown in Fig. 1A, IHC illustrated that the expression of UPF1 was decreased in HCC tissues compared with
adjacent non-tumor tissues. We next detected the expression of UPF1 in these tissues by real time PCR and also found the low expression of UPF1 in HCC tissues (Fig. 1B). Fifty cases were
divided into two groups: a high UPF1 expression group (above the median UPF1 expression) and a low UPF1 expression group (below the median). As shown in Supplementary Table S1, the
correlation regression analysis revealed that the expression of UPF1 was associated with tumor size and lymph node metastasis. These indicated that UPF1 was involved in HCC and might play an
important role in tumor progression. SILENCING OF UPF1 PROMOTED HCC CELLS GROWTH AND INVASION To further investigate the roles of UPF1 in HCC, we silenced UPF1 expression by transfecting
specific siRNAs targeting UPF1 into HCC Huh7 and HepG2 cells. The transfection efficiently decreased the expression of UPF1 valuated by western blotting (Fig. 2A,E). Among three siRNAs the
second and third ones were more efficient, and therefore the number #2 and #3 were used to conduct the subsequent experiments. Firstly, MTT assay was carried out in HCC cell lines. After
expression of UPF1 was downregulated in Huh7 and HepG2 cells, the cell proliferation increased (Fig. 2B,F). Colony formation assay was also used to test effect of UPF1 on cell growth. Figure
2C,G showed that the colony-forming ability of cells was enhanced after knockdown of UPF1. We next assessed the effects of UPF1 on cell invasion in Huh7 and HepG2 cells. As shown in Fig.
2D,H, knockdown of UPF1 significantly increased cell invasion rates, moreover, the overexpression of UPF1 decreased cell invasion ability. These findings suggested that silencing of UPF1
promoted HCC cells growth and invasion. UPF1 COULD BIND TO LONG NON-CODING RNA UCA1 Recently, there are several reports that many lncRNAs participate in molecular regulation pathways through
their interactions with proteins15. Online software starBase v2.0 database was used to identify lncRNAs that potentially combine with UPF1. We found that UCA1 was able to bind to UPF1.
Firstly, the expression level of UCA1 was studied in HCC by real time PCR. Results showed that the expression of UCA1 was higher in HCC than adjacent noncancerous HCC tissues (Fig. 3A). To
further explore the relationship between UPF1 and UCA1 in HCC, we analyzed their expression levels in HCC tissues. Figure 3B showed that the RNA expression level of UPF1 and UCA1 in the
paired tissues was negative associated by RT-PCR. RNA stability of UCA1 was further tested in HCC cells with UPF1 knockdown. As shown in Fig. 3C, the decay rate of UCA1 was increased in Huh7
after knockdown of UPF1. Next, we analyze the binding between UCA1 and UPF1 by RIP, and found that UPF1 specially combined with UCA1 (Fig. 3D). We also transfected UPF1 expressed plasmid
into Huh7 cells. The overexpression of UPF1 decreased the expression of UCA1 (Fig. 3E). While, the knockdown of UPF1 in Huh7 cells increased the expression of UCA1 (Fig. 3F). The above data
hinted that UPF1 combined with UCA1 and could potentially take part in HCC progression. EFFECT OF UCA1 EXPRESSION ON HCC GROWTH AND INVASION We also analyzed the correlation between UCA1
expression and clinicopathological characteristics of these 50 HCC patients. Fifty cases were divided into two groups: a high UCA1 expression group (above the median UCA1 expression) and a
low UCA1 expression group (below the median). As shown in Supplementary Table S2, the correlation regression analysis revealed that high expression of UCA1 was associated with large tumor
size (_p_ = 0.032) and lymph node metastasis (_p_ = 0.033), which implied that UCA1 may be involved in the growth and invasion of HCC. Then the expression of UCA1 in Huh7 and HepG2 cells was
knocked down to study the function of UCA1 and the siRNA number #2 and #3 were more efficient target for UCA1 to conduct subsequent experiments (Fig. 4A,E). To examine the effects of UPF1
on HCC growth, MTT assay have been done. As shown in Fig. 4B,F, knockdown of UCA1 decreased the cell progression of HCC cells. In addition, colony formation assay showed that the knockdown
of UCA1 reduced the Huh7 and HepG2 cell growth (Fig. 4C,G). We next evaluated the effects of UCA1 on cell invasion by transwell assay. After down-regulation of UCA1 expression, the invasion
capacity was decreased, and then overexpression of UCA1 increased cell invasion (Fig. 4D,H). All these data suggested that UCA1 may be involved in HCC growth and invasion. KNOCKDOWN OF UCA1
AMELIORATED THE EFFECT OF UPF1 KNOCK DOWN ON HCC GROWTH AND INVASION To further investigate the relationship between UPF1 and UCA1, the siRNA for UCA1 was transfected into HCC after
knockdown of UPF1. Firstly, real time PCR showed that the knockdown of UCA1 decreased the expression of UCA1 after UPF1 knock down increasing UCA1 (Fig. 5A). MTT assay showed that the
knockdown of UPF1 increased the HCC proliferation, while UCA1 knock down decreased the cell proliferation ability (Fig. 5B). Next colony formation assay was used to further study cell
growth, which was also weakened after the knockdown of UCA1 (Fig. 5C). Finally, we observed that cell invasion ability was decreased after the knockdown of UCA1 by transwell assay (Fig. 5D).
These suggested that knockdown of UCA1 ameliorated the effect of UPF1 knock down on HCC growth and invasion. KNOCKDOWN OF UPF1 ENHANCES GLYCOLYSIS IN HCC Usually tumor cells exhibit a
higher rate of glucose metabolism compared with normal cells16. It is reported that UCA1 was involved in tumor cell glycolysis17. Next, we investigated the changes in glycolysis in HCC with
knockdown of UPF1. The results showed that knockdown of UPF1 increased the rates of glucose consumption in HCC (Fig. 6A). We also found that UPF1 knockdown increased the rates of lactate
production in HCC (Fig. 6B). These suggested that knockdown of UPF1 enhances glycolysis in HCC. DISCUSSION Here, we reported that the correlation between UPF1 and HCC. IHC and real time PCR
showed that UPF1 expression was decreased in HCC and might play an important role in tumor progression. We also identified the function of UPF1 in the HCC cells by applying loss-of-function
approaches. Our data clearly demonstrated that knockdown of UPF1 promoted HCC cells growth and invasion. Up-frameshift (Upf) complex facilitates the degradation of aberrant mRNAs18,19. UPF1
has been characterized as an essential factor for nonsense-mediated mRNA decay (NMD)20,21. UPF1 is essential for embryonic development and survival22,23. The loss of UPF1 function inhibits
cell growth and induces apoptosis in Drosophila melanogaster24. UPF1 also takes part in cancer progression. UPF1 is a potential modulator of MALAT1 and that UPF1/MALAT1 pathway could be a
therapeutic target for gastric cancer25. UPF1 was expressed at lower levels in human lung adenocarcinoma tissues than in normal lung tissues, thereby raising the possibility that NMD may be
downregulated to permit lung adenocarcinoma oncogenesis26. Our results were consistent with these previous reports that UPF1 was a tumor suppressor. Long non-coding RNAs (lncRNAs) are
defined as those >200 nucleotides (nt). LncRNAs are coming from the “noisy region” of the genome as a new source of biomarkers that could characterize disease recurrence and
progression27. LncRNAs function in various aspects of cell biology has focused increasing attention on their potential to contribute towards tumor development28,29,30. Urothelial cancer
associated 1 (UCA1) is a long noncoding RNA having aberrant expression in embryogenesis and a broad range of cancer tissues and cells31,32. Especially, UCA1 plays oncogenic roles in tumor
growth and metastasis33,34. However, the molecular mechanism of UCA1 in cancer progression remains incompletely understood. In this study, UPF1 could bind to UCA1 and the correlation between
UPF1 and UCA1 was negative. Knockdown of cellular UCA1 significantly decreased the cell growth and invasion. In conclusion, our present study uncovers a novel UPF1-mediated mechanism of
cell growth and invasion by targeting long non-coding RNA UCA1 in HCC cells. Our results indicated that UPF1/UCA1 played an important role during HCC tumorigenesis and may serve as a
putative target for HCC diagnosis and therapy. REFERENCES * Venook, A. P., Papandreou, C., Furuse, J. & de Guevara, L. L. The incidence and epidemiology of hepatocellular carcinoma: A
global and regional perspective. _Oncologist._ 15(Suppl 4), 5–13 (2010). Article Google Scholar * Chrostek, L. & Panasiuk, A. Liver fibrosis markers in alcoholic liver disease. _World
J Gastroenterol._ 20, 8018–8023 (2014). Article Google Scholar * Fujiwara, N., Friedman, S. L., Goossens, N. & Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in
the era of precision medicine. _J Hepatol._ 68, 526–549 (2018). Article Google Scholar * Ali, E. S., Rychkov, G. Y. & Barritt, G. J. Metabolic Disorders and Cancer: Hepatocyte
Store-Operated Ca2+ Channels in Nonalcoholic Fatty Liver Disease. _Adv Exp Med Biol._ 993, 595–621 (2017). Article CAS Google Scholar * Du, J. _et al_. Hepatitis B core protein promotes
liver cancer metastasis through miR-382-5p/DLC-1 axis. _Biochim Biophys Acta._ 1865, 1–11 (2017). Article ADS Google Scholar * Gosalia, A. J., Martin, P. & Jones, P. D. Advances and
Future Directions in the Treatment of Hepatocellular Carcinoma. _Gastroenterol Hepatol (N Y)._ 13, 398–410 (2017). Google Scholar * Riordan, J. D. _et al_. Chronic liver injury alters
driver mutation profiles in hepatocellular carcinoma. _Hepatology._ 67, 924–939 (2018). Article CAS Google Scholar * Yarchoan, M. _et al_. Characterization of the Immune Microenvironment
in Hepatocellular Carcinoma (HCC). _Clin Cancer Res._ 23, 7333–7339 (2017). Article CAS Google Scholar * Applequist, S. E., Selg, M., Raman, C. & Jäck, H. M. Cloning and
characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. _Nucleic Acids Res._ 25, 814–821 (1997). Article CAS Google Scholar *
Imamachi, N., Tani, H. & Akimitsu, N. Up-frameshift protein 1 (UPF1): multitalented entertainer in RNA decay. _Drug Discov Ther._ 6, 55–61 (2012). CAS PubMed Google Scholar * Lou, C.
H. _et al_. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. _Cell Rep._ 6, 748–764 (2014). Article CAS Google Scholar *
Chang, L. _et al_. The human RNA surveillance factor UPF1 regulates tumorigenesis by targeting Smad7 in hepatocellular carcinoma. _J Exp Clin Cancer Res._ 35, 8 (2016). Article Google
Scholar * Zhou, Y., Li, Y., Zheng, J., Liu, K. & Zhang, H. Detecting of gastric cancer by Bcl-2 and Ki67. _Int J Clin Exp Pathol._ 8, 7287–7290 (2015). CAS PubMed PubMed Central
Google Scholar * Kotake, Y. _et al_. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. _Oncogene._ 30, 1956–1962 (2011).
Article CAS Google Scholar * Yang, F. _et al_. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. _Mol Cell._ 49, 1083–1096
(2013). Article CAS Google Scholar * Birsoy, K. _et al_. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. _Nature._ 508, 108–112 (2014). Article
ADS CAS Google Scholar * Li, Z., Li, X., Wu, S., Xue, M. & Chen, W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143
pathway. _Cancer Sci._ 105, 951–955 (2014). Article CAS Google Scholar * Kurosaki, T. & Maquat, L. E. Nonsense-mediated mRNA decay in humans at a glance. _J Cell Sci._ 129, 461–467
(2016). Article CAS Google Scholar * Lee, S. R., Pratt, G. A., Martinez, F. J., Yeo, G. W. & Lykke-Andersen, J. Target Discrimination in Nonsense-Mediated mRNA Decay Requires Upf1
ATPase Activity. _Mol Cell._ 59, 413–425 (2015). Article CAS Google Scholar * He, F. & Jacobson, A. Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of
the Story. _Annu Rev Genet._ 49, 339–366 (2015). Article CAS Google Scholar * Fiorini, F., Bagchi, D., Le Hir, H. & Croquette, V. Human Upf1 is a highly processive RNA helicase and
translocase with RNP remodelling activities. _Nat Commun._ 6, 7581 (2015). Article Google Scholar * Wittkopp, N. _et al_. Nonsense-mediated mRNA decay effectors are essential for zebrafish
embryonic development and survival. _Mol Cell Biol._ 29, 3517–3528 (2009). Article CAS Google Scholar * Medghalchi, S. M. _et al_. Rent1, a trans-effector of nonsense-mediated mRNA
decay, is essential for mammalian embryonic viability. _Hum Mol Genet._ 10, 99–105 (2001). Article CAS Google Scholar * Avery, P. _et al_. Drosophila Upf1 and Upf2 loss of function
inhibits cell growth and causes animal death in a Upf3-independent manner. _RNA._ 17, 624–638 (2001). Article Google Scholar * Li, L. _et al_. The Human RNA Surveillance Factor UPF1
Modulates Gastric Cancer Progression by Targeting Long Non-Coding RNA MALAT1. _Cell Physiol Biochem._ 42, 2194–2206 (2017). Article CAS Google Scholar * Cao, L. _et al_. Human
nonsense-mediated RNA decay regulates EMT by targeting the TGF-ß signaling pathway in lung adenocarcinoma. _Cancer Lett._ 403, 246–259 (2017). Article CAS Google Scholar * Han, D. _et
al_. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. _Oncotarget._ 7, 22159–22173 (2016). PubMed PubMed
Central Google Scholar * Bhan, A., Soleimani, M. & Mandal, S. S. Long Noncoding RNA and Cancer: A New Paradigm. _Cancer Res._ 77, 3965–3981 (2017). Article CAS Google Scholar *
Bolha, L., Ravnik-Glavač, M. & Glavač, D. Long Noncoding RNAs as Biomarkers in Cancer. _Dis Markers._ 2017, 7243968 (2017). Article Google Scholar * Wu, H., Yang, L. & Chen, L. L.
The Diversity of Long Noncoding RNAs and Their Generation. _Trends Genet._ 33, 540–552 (2017). Article CAS Google Scholar * Xue, M., Chen, W. & Li, X. Urothelial cancer associated 1:
a long noncoding RNA with a crucial role in cancer. _J Cancer Res Clin Oncol._ 142, 1407–1419 (2016). Article Google Scholar * Wang, Z. Q. _et al_. Long noncoding RNA UCA1 promotes tumour
metastasis by inducing GRK2 degradation in gastric cancer. _Cancer Lett._ 408, 10–21 (2017). Article CAS Google Scholar * Wang, Z. Q. _et al_. Long noncoding RNA UCA1 induced by SP1
promotes cell proliferation via recruiting EZH2 and activating AKT pathway in gastric cancer. _Cell Death Dis._ 8, e2839 (2017). Article CAS Google Scholar * Zhang, S., Dong, X., Ji, T.,
Chen, G. & Shan, L. Long non-coding RNA UCA1 promotes cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer. _Am J Transl Res._ 9, 366–375 (2017). CAS
PubMed PubMed Central Google Scholar Download references AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Gastroenterology, The First Affiliated Hospital of Xi’an Medical
University, Xi’an, China Yongli Zhou & Liqiao Ge * Department of Pathology, The First Affiliated Hospital of Xi’an Medical Universtiy, Xi’an, China Yandong Li & Jianyun Zheng *
General Medical College, Xi’an Medical University, Xi’an, China Yongli Zhou, Yandong Li, Jianyun Zheng & Liqiao Ge * Department of Hematology, General Hospital of North China Petroleum
Administration Bureau, Renqiu, China Na Wang * Shanxi University of Traditional Chinese Medicine, Taiyuan, China Xiuying Li Authors * Yongli Zhou View author publications You can also search
for this author inPubMed Google Scholar * Yandong Li View author publications You can also search for this author inPubMed Google Scholar * Na Wang View author publications You can also
search for this author inPubMed Google Scholar * Xiuying Li View author publications You can also search for this author inPubMed Google Scholar * Jianyun Zheng View author publications You
can also search for this author inPubMed Google Scholar * Liqiao Ge View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Li Y. conceived and
designed this study. Zhou Y., Wang N., Li X., Zheng J. and Ge Q. performed and analysed the experiments. Zhou Y. and Li Y. wrote this paper. All authors read and approved the manuscript.
CORRESPONDING AUTHOR Correspondence to Yongli Zhou. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer
Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION RIGHTS AND PERMISSIONS OPEN
ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format,
as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third
party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the
article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright
holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Zhou, Y., Li, Y., Wang, N. _et al._
UPF1 inhibits the hepatocellular carcinoma progression by targeting long non-coding RNA UCA1. _Sci Rep_ 9, 6652 (2019). https://doi.org/10.1038/s41598-019-43148-z Download citation *
Received: 16 August 2018 * Accepted: 05 March 2019 * Published: 30 April 2019 * DOI: https://doi.org/10.1038/s41598-019-43148-z SHARE THIS ARTICLE Anyone you share the following link with
will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative