An algorithm for enhancing the image contrast of electron tomography
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Three-dimensional (3D) reconstruction of a single protein molecule is essential for understanding the relationship between the structural dynamics and functions of the protein.
Electron tomography (ET) provides a tool for imaging an individual particle of protein from a series of tilted angles. Individual-particle electron tomography (IPET) provides an approach for
reconstructing a 3D density map from a single targeted protein particle (without averaging from different particles of this type of protein), in which the target particle was imaged from a
series of tilting angles. However, owing to radiation damage limitations, low-dose images (high noise, and low image contrast) are often challenging to be aligned for 3D reconstruction at
intermediate resolution (1–3 nm). Here, we propose a computational method to enhance the image contrast, without increasing any experimental dose, for IPET 3D reconstruction. Using an
edge-preserving smoothing-based multi-scale image decomposition algorithm, this method can detect the object against a high-noise background and enhance the object image contrast without
increasing the noise level or significantly decreasing the image resolution. The method was validated by using both negative staining (NS) ET and cryo-ET images. The successful 3D
reconstruction of a small molecule (<100 kDa) indicated that this method can be used as a supporting tool to current ET 3D reconstruction methods for studying protein dynamics via
structure determination from each individual particle of the same type of protein. SIMILAR CONTENT BEING VIEWED BY OTHERS LOTTOR: AN ALGORITHM FOR MISSING-WEDGE CORRECTION OF THE LOW-TILT
TOMOGRAPHIC 3D RECONSTRUCTION OF A SINGLE-MOLECULE STRUCTURE Article Open access 26 June 2020 SINGLE-PARTICLE CRYO-EM AT ATOMIC RESOLUTION Article 21 October 2020 HIGH-RESOLUTION IN SITU
STRUCTURE DETERMINATION BY CRYO-ELECTRON TOMOGRAPHY AND SUBTOMOGRAM AVERAGING USING EMCLARITY Article 12 January 2022 INTRODUCTION In solution, protein particles travel in following the
Brownian motion and their structures are vibrated in following the thermodynamics. Studying the dynamics of a type of protein requires a capability for 3D structure determination of an
individual particle of this type of protein. Conventional structure determination approaches, including X-ray crystallography and cryo-electron microscopy (cryo-EM) single-particle 3D
reconstruction, require an averaging process on thousands to millions of different particles of the same type of protein1. Averaging the structures from different particles without prior
knowledge of their structure identity, or ignored the nature flexibility or fluctuation of the particles may lead to artifacts in protein structure determination such as blurring or
eliminating the flexible domains. The typical flexble proteins include the double strand DNA (dsDNA)2,3,4,5, antibody6,7,8, lipoprotein and neuron proteins9,10,11,12,13,14. As an example of
the IgG1 antibody, by enforced averaging the particle images of IgG antibody in single-particle 3D reconstruction6,7,8, one or two domains could disappeared. An ideal approach to reveal the
protein dynamics is to tracking the 3D structure changes on each individual particle of protein in liquid solution. Since no technique is available for imaging the 3D structure of an
individual particle of protein in solution, the molecular dynamic (MD) simulation was often used to reveal the dynamics of an individual particle of protein in solution. An alternative
approach to reveal the protein 3D structural dynamics is to determine the 3D structures of hundreds of individual particles that were frozen or fixed in a same time. The statistical analysis
of the variety of the 3D structures from different particles can reflect the 3D dynamics of this type of protein. Electron tomography (ET) is a powerful tool for imaging a targeted particle
from a series of tilted viewing angles. The computerized image algorithms enable us to align the images and reconstruct them into a 3D density map. The 3D reconstruction resolution
significantly depends on the accuracy of the alignment of the tilted images and the noise levels of the images. High noise level can decrease the accuracy of the image alignment.
Unfortunately, in the experiment, the capability to reduce the noise level of the images was limited by the radiation damage and the dose tolerance limitation. To decrease the noise level
(increase the image contrast) without increasing the imaging dose or radiation damage, a computational approach was here introduced. The approach included edge-preserving smoothing filters
proposed by L.P. Yaroslavsky in 198515, as an image-optimizing tool widely used in computer vision. By smoothing images while effectively preserving edges, this method can enhance image
contrast by decomposing an image into piecewise smooth layers and detailed layers15. In 1990, Perona and Malik adopted a diffusion process to realize semantically meaningful edges16. To
further improve edge-smoothing capabilities, two non-linear Gaussian-filter-based edge-preserving methods were introduced by Aurich and Weule in 199517 and by Smith and Brady in 199718.
Since then, more image decomposition methods have been reported based on those edge-preserving smoothing models19. In most cases, a combination of multi-scale operations was used20. The
limitations of these methods include some halo artifacts existing near the boundaries from using previous models21. To overcome these limitations, several optimized edge-preserving smoothing
filters have been reported16,22,23,24. Among these methods, the bilateral filter (BLF)23 and weighted least squares (WLS) filter22 are two of the most effective tools for multi-scale
decomposition, especially for detail extraction and noise removal. Compared with the BLF filter, the WLS filter is more convenient, flexible and appropriate for extracting multi-scale
details. Here, we introduce a method to enhance the image contrast of ET by using this WLS filter as an edge-preserving smoothing filter. The model was more effective than the traditional
edge-preserving decomposition-based detail manipulation method22. To validate the capability, we tested this method to enhance the low-contrast simulated images, real experimental cryo-EM
and cryo-ET images, and real experimental negative staining (NS) ET images, as well as the real experimental cryo-positive staining (PS) ET images. The final 3D reconstructions showed that
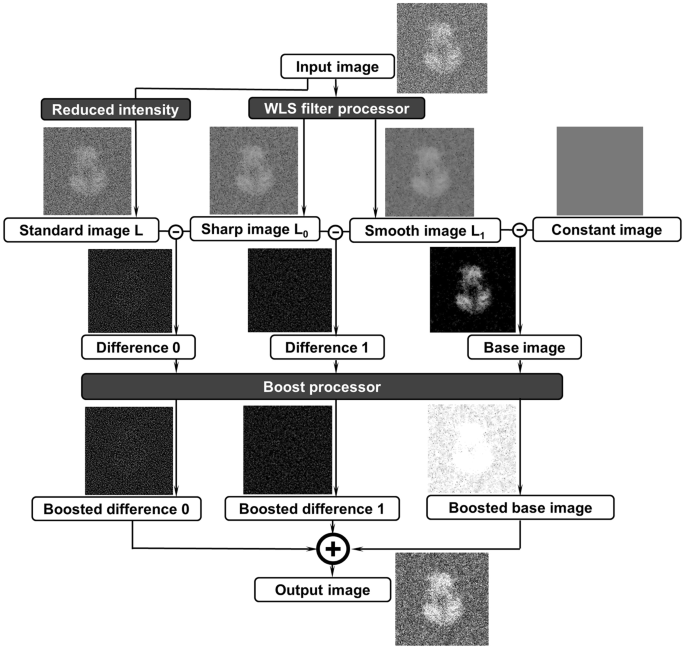
this method can be useful for the 3D structure determination of an individual particle of protein by ET. OVERVIEW OF THE ALGORITHM Flow chart of the algorithm of the edge-preserving
smoothing-based multi-scale image decomposition for image contrast enhancement showed in Fig. 1 contains two processes, the WLS filter and image boosting. In the process of WLS filter, the
input image was decomposed into 3 different images, standard image L, relatively sharp image L0 and relatively smooth image L1. L is an optimized image after reduced the intensity for 3
times on the input image. L0 and L1 are two processed images filtered by sharp-scale and common-scale WLS filters, respectively. Constant image was a blank image with a magic intensity of
56, which reflects the image’s background information. By subtracting from each other, three difference images, difference 0, difference 1 and base image were submitted for a boosting
process for extracting the original image detail and base information at different dimensions. By combining those three boosted images into one, the output image can capture the major
details in the different levels while maintaining the image’s base information. In the process, the WLS filter processer and boost processor as essential models can directly contribute to
enhancing the image contrast, as presented in details in the following sections. THE PROCESS OF THE WLS FILTER Compared with other edge-preserving smoothing filters, the WLS filter22 has
obvious advantages, especially for multi-scale decomposition and detail extraction. Through a WLS filter with appropriate parameters, a coarse, piecewise smooth version of decomposition can
be obtained. Moreover, a sequence of images capturing different details at progressively finer scales is presented. As shown in Fig. 1, image _L_0 is obtained by using a sharp-scale WLS
filter, and image _L_1 is obtained by using a common-scale WLS filter. Image _L_0 retains more details than image _L_1 (significant gradients). The constant image presents the background of
the image with a constant intensity of 56 as a magic number used for any images. The differences between images _L_, _L_0 and _L_1 reflect the image details at different levels. THE PROCESS
OF IMAGE BOOSTING As discussed above, the difference images reflect the image details at different levels (Fig. 1). However, we found that by directly calculating the differences, it was
difficult to control the degree of detail in the layers. In some cases, the detailed layers were either significantly boosted or not obvious. Thus, this strategy to control the levels of
detailed layers was challenging. In this case, we proposed an effective boosting function25,26 as follows: $$y=\frac{1}{1+{e}^{-ax}}-0.5$$ (1) where _x_ indicates one input processed image,
and _a_ indicates the sigmoid parameter that determines the degrees of sigmoid. In our case, _a_ = 8 was used. The boosting function above can be considered a sigmoid curve that can shift
and normalize the target term. It not only contributed to controlling the contrast and exposure of the base layer but also contributed to keeping the boosting of details under controlling. A
rescale process can be added later if necessary for the experimental data. The intermediate states of images of an example were used to show the changes of images before and after boosted
process (Fig. 1). We next applied the algorithm to both simulated and experimental images to test how the edge-preserving smoothing-based image decomposition models can effectively enhance
the image contrast, in which some images were further evaluated by the 3D reconstruction. IMPLEMENTATION ON THE LOW-CONTRAST 2D IMAGES The enhancement algorithm was first tested on simulated
2D low-dose cryo-EM images. Considering the smaller protein is more challenging for imaging and 3D reconstruction, two proteins with different molecule weight were used: (i) a fragment (A-D
chains, molecular mass: ~108 kDa) of a molybdate transporter (ModB2C2, PDB accession number 2ONK27) and (ii) cholesteryl ester transfer protein (CETP, PDB accession number 2OBD28, molecular
mass: ~53 kDa). The simulated images of these two proteins were prepared by using the following protocol: (a) The simulated 3D density maps were generated by using the “_pdb2mrc_” command
of the EMAN software package29, in which the map of the ModB2C2 protein was generated at a resolution of 4 Å within a box of 160 × 160 × 160 voxels (voxel size of 1 Å), and the map of CETP
was generated at a resolution of 2 Å within a box of 192 × 192 × 192 voxels (voxel size of 1 Å). (b) The projection images of the maps were generated by using the “_PJ 3Q_” command of the
SPIDER software package30. In the projections, each pixel was 1 Å. (c) To simulate noise in the cryo-EM image for evaluating the effect of enhancement method on images with different noise
levels, Gaussian-type noises were applied to the above projections to achieve a final SNR of 0.80, 0.50 and 0.30 respectively. For ModB2C2, the Fourier ring correlation (FRC) curves
calculated between the enhanced image and noise-free original reference image were similar to the FRC curves calculated between the non-enhanced images and noise-free original reference
image (Fig. 2A). This result suggests that, although the enhancement method did not significantly change the image resolution, the image contrast was still significantly enhanced. The SNR
levels increased by 60–70% compared with non-enhanced images, from 0.80 to 1.34, from 0.50 to 0.80 and from 0.30 to 0.53 (Table 1). Similar results were obtained for the small protein, CETP.
The analyses showed that the FRC curves were also similar to the curves before applied the enhancement (Fig. 2B). The SNR levels were also increased by 60–70%, from 0.80 to 1.33, from 0.50
to 0.83 and from 0.30 to 0.49 (Table 2). The enhancement algorithm was also tested on experimental cryo-EM images of Termoplasma acidophilum 20S proteasome31 (Supplemental Fig. S1A,
downloaded from Electron Microscopy Public Image Archive, entry EMPIAR-10025). These cryo-EM images were acquired on FEI Titan Krios TEM equipped with a Gatan K2 Summit direct detector,
which has been used to achieve a 3D single-particle reconstruction at 2.8 Å resolution31. In the raw images, proteasome particles were barely visible (Supplemental Fig. S1A). After low-pass
filtering the images at 8 Å, the particles became clear (Supplemental Fig. S1B), allowing us to easily box the particles from the images. One representative boxed particle showed a boost of
SNR from ~0.06 to ~0.55 by the low-pass filtering (Supplemental Fig. S1C,D). The SNR of low-pass filtered images could be further increased to ~0.72 (~30% increase) by the enhancement
algorithm (Supplemental Fig. S1E). The FRC curve between the raw image and the image after enhancement (also with low-pass filtered at 8 Å) is higher than 0.9 at frequency higher than 1/4
Å−1 (Supplemental Fig. S1F), demonstrating that the resolution of image was maintained by the enhancement algorithm. This result also evidenced that, although the density of image after
enhancement does not reflect electron density of protein any more (due to the nonlinear boosting function of the enhancement), the information about protein structure was reserved in the
enhanced images. The above tests using both simulated and experimental 2D images showed that the enhancement procedure had no significant effect on the resolution of the images but increased
the image contrast. The effect of contrast enhancement would not be reduced as the particle size decreased. Thus, the enhancement method can be used to improve the image constrast of 2D
images of particles in various sizes. IMPLEMENTATION ON THE SIMULATED 3D RECONSTRUCTION TESTING THE EFFECT OF THE ENHANCEMENT METHOD ON THE SIMULATED 3D RECONSTRUCTION It is well known that
the 3D reconstruction resolution was significantly depended on two things, the noise level (image contrast) of the 2D tilt series and the accuracy of the alignment of those 2D tilt series.
The noise can directly reduce the 3D reconstruction resolution and indirectly influence the accuracy of the alignment to further reduce the 3D reconstruction resolution. To improve the 3D
reconstruction via increasing the accuracy of the alignment of the 2D tilt series, we conducted following processes on simulated data. A set of simulated tilt series (perfectly aligned,
without image shift) was generated by projecting ModB2C2 from a tilt angle range of −90° to +90° in steps of 1° by using the “_PJ 3Q_” command (SPIDER software package)30. Gaussian noise was
then added to the tilt series in SNRs of 0.80, 0.50, and 0.30 respectively. After contrast enhancement, each tilt series were directly back projected into a 3D reconstruction by using the
“_BP 3F_” command (SPIDER software package). The Fourier shell correlation (FSC) curves computed between the 3D reconstruction and reference 3D object (noise free) showed that the
enhancement method did not significantly change the 3D resolution nor change the structure (Fig. 3A); however, it did increase the SNR of the 3D reconstruction by approximately 50% (from SNR
of 0.70 to 1.09, 0.42 to 0.66 and 0.27 to 0.43 respectively) (Table 1). Similarly, by using a small protein, ~53 kDa CETP, 3D reconstructions were evaluated under the tilt series SNR levels
of 0.80, 0.50, and 0.30 respectively. The FSC analyses of the 3D reconstructions showed no significant change in resolution (Fig. 3B), but the SNR levels were increased by 50%, (from 0.61
to 0.94, 0.38 to 0.61, and 0.24 to 0.37) (Table 2), which was consistent to the larger protein (ModB2C2). The above evaluations showed that the enhancement method did not decrease the 3D
reconstruction resolution and the reconstructed 3D map has similar features to that without enhancement, except few tiny differences (Fig. 3). However, the enhancement method improved the
contrast of 3D reconstruction by ~50%. A further improvement in 3D constrast could be obtained by prior applied the low-pass filter before the enhancement. To demonstrate this approach, the
images of ~108 kDa ModB2C2 samples were generated with super low SNRs, 0.25, 0.20 and 0.15, respectively. Each image was filtered by the low-pass filter at ~8 Å and then submitted to the
enhancement. Statistical analyses showed that the contrast of 2D images were significantly improved after low-pass filterring as expected, _i.e_., the SNRs of the 2D images were increased
from 1.49, 0.9 and 0.81 to 2.27, 1.31 and 1.14 respectively (Table 1). More importantly, the 3D reconstructions showed that the enhancement method led to less than 2% decreasing in the
resolution (from ~7.9 Å, ~8.8 Å and ~10.8 Å to ~8.0 Å, ~8.9 Å and ~10.9 Å, respectively), but increased the 3D SNRs from 4.06, 3.27 and 2.54 to 4.64, 3.89 and 3.09 respectively (Table 1).
The similar results were also obtained on small protein, CETP (Table 2). The SNRs of the CETP images at 0.25, 0.2 and 0.15 were increased to 1.57, 1.25, and 0.93 by low-pass filtering at ~8
Å, and then further increased to 2.28, 1.76, and 1.26 by the enhancement method. SNRs of the 3D reconstructions of CETP were increased from 3.96, 3.15, and 2.39 by low-pass filterring, and
then further increased to 4.89, 3.88 and 2.90 respectively. The 3D resolutions were changed within a range of ~5%, _i.e_., from ~4.42 Å, ~5.75 Å, and ~7.73 Å to ~4.64 Å, ~5.95 Å and ~7.79 Å
respectively. Therefore, the combination of low-pass filter and enhancement can benefit the 3D reconstructions of super high noisy images, such as the low-dose cryo-ET images. EVALUATING THE
ENHANCEMENT ON IPET 3D RECONSTRUCTION OF SIMULATED ELECTRON TOMOGRAPHY The resolution of 3D reconstruction was depended on both the contrast of 2D tilt images and the accuracy of the image
alignment. To evaluate the accuracy of the alignment of the enhanced images in IPET 3D reconstruction of an individual particle of protein (no averaging on different particles of same type
of protein), we applied the method to a set of simulated tilt series of ET images of a single molecule, a ModB2C2 molecule. In this tilt series, the center of each tilt image was randomly
shifted away from the center within a range of 30 pixels to simulate the translational errors. The enhancement process was applied to the simulated tilt series with SNR = 0.3 before IPET 3D
reconstruction1. The process of IPET 3D reconstruction (Fig. 4A) showed that, as the alignment of the enhanced images was gradually improved, the contrast of the 3D reconstruction was
gradually enhanced. The final 3D reconstruction showed the features of the secondary structure, such as the α-helix. To further test the effect of the enhancement method on the IPET 3D
reconstruction of small protein, CETP, the tilt series with SNR = 0.30 and center-shifted range of 30 pixels was generated, then submitted for enhancement before conducting the IPET 3D
reconstruction (Fig. 4B). The process showed that the tilt images were systematically enhanced and aligned. The final 3D reconstruction confirmed that the high-resolution details could be
reconstructed after the enhancement (Fig. 4B). The above evaluations showed that images after enhancement could be well aligned for achieving a 3D reconstruction without losing the high
resolution structure details, even for a small protein like CETP under a high noise condition. The results suggested that the enhancement method did not bias the image alignment thus can be
used to improve the quality (SNR) of IPET 3D reconstruction of a single molecule. IMPLEMENTATION ON EXPERIMENTAL 3D RECONSTRUCTION EVALUATING THE ENHANCEMENT ON THE IPET 3D RECONSTRUCTION OF
NEGATIVE-STAINING ELECTRON TOMOGRAPHY After confirmed the enhancement method regarding that it did not decrease the resolution of 3D reconstruction but increase the SNR on simulation data,
we applied the method to the real experimental data. In this section, we applied the method to a set of high contrast negative staining tilt series of an 84-base-pair double-stranded DNA
(dsDNA, with a molecular mass of ~52 kDa) conjugated to 5 nm nanogold. In the next section, we applied the method to a set of low contrast tilt series, _i.e_., a set of cryo-EM tilt series
of a particle of LDL bound to CETP32 (the molecular mass of LDL and CETP are ~2,500 kDa33 and ~53 kDa, respectively), and to a set of cryo-PS tilt series of a particle of DNA origami
acquired by using direct detector. The high contrast tilt series of a dsDNA-nanogold conjugate was imaged from −60° to +60° in steps of 1.5° 3. The three representative survey views showed
each fiber-shaped dsDNA bound to two nanogold particles from each of its distal ends (Fig. 5A). Before the targeted particle was tracked and boxed from the tilt series, the CTF of the tilt
series was measured and corrected by TomoCTF software package34. The single complex of dsDNA-nanogold conjugates was previously published; thus, the double complexes of the dsDNA-nanogold
conjugates were used here for 3D reconstruction. The selected tilt view of the double complexes showed that the dsDNA portions were scarcely visible (left column in Fig. 5B) and the SNRs
were within the range of ~0.17 to ~0.70 with an average of ~0.46. By the enhancement process, the SNRs was slightly increased to the range from ~0.22 to ~0.85 with an average of ~0.56, and
the overall shape of the dsDNA became slightly visible (Fig. 5B, left second column). Through the IPET 3D reconstruction, the final 3D image showed two handcuff-shaped complexes attached to
each other (Fig. 5C). The FSC analysis showed the 3D map had a resolution of ~17.6 Å (Fig. 5E). The overall conformation of each complex was similar to that previously published3. However,
two complexes seemed attached to each other, owing to the interaction of irregularly shaped densities coating the surfaces of the nanogold particles (Fig. 5C). These densities may possibly
have been thiolated short-chain polyethylene glycol (PEG) molecules that were used to stabilize the particles against aggregation at high ionic strength35. To reveal the approximate
conformation of the dsDNAs, the standard model of 84-base-pair dsDNA was manually and flexibly docked into the fiber-shaped density between two nanogold particles in each complex (Fig. 5D).
Because of the opposite image contrast of the nanogold particles relative to that of the DNA, we reversed the image contrast of the final 3D image (colored in gold) and overlaid this 3D
image over the original 3D image to display both the DNA and nanogold particles in the same 3D map (Fig. 5D). This overlaid map showed nanogold particles with diameters of ~79.0 Å, ~59.0 Å,
~56.0 Å and ~65.0 Å. Each of the two high-density fabrics with overall dimensions of ~240.0 Å long and ~15 to ~25 Å wide bridged two nanogolds (Fig. 5D). Similarly, another 3D density map of
double complexes of dsDNA-nanogold conjugates was reconstructed by combining the enhancement method and IPET (Fig. 5F–I). The enhancement method slightly increased the SNRs of each tilt
image from ~0.10 to ~0.47 (with an average of 0.27) to ~0.14 to ~0.54 (with an average of ~0.31). The 3D reconstruction showed that two handcuff-shaped particles were connected to each other
by the surface interaction of two nanogolds in each complex. The ~17 Å resolution was measured on the basis of FSC analyses (Fig. 5I); the conformation was similar to that of the first
double complexes. The overlaid density map from the final 3D density map and its reversed density map (colored in gold) showed that the nanogold particles had diameters of ~54.0 Å, ~63.0 Å,
~58.0 Å and ~60.0 Å, and were bridged by two fabric-like DNA densities with overall dimensions of ~255 to ~275 Å long and ~15 to 25 Å wide (Fig. 5H). The conformations of dsDNA strands were
obtained by manually and flexibly docking the standard structure of 84-base-pair dsDNA models into the bridging portion density (Fig. 5H). Successfully repeating the previous IPET 3D
reconstruction and achieving 3D reconstructions of the double complexes from negative-staining ET data confirmed that the enhancement method is a reliable method to preprocess tilt images
before IPET 3D reconstruction. The results obtained from the above process demonstrated a successful case in which the enhancement method benefits the IPET 3D reconstruction of a single
target object based on the low contrast negative-staining images. EVALUATING THE ENHANCEMENT ON THE IPET 3D RECONSTRUCTION OF CRYO-ELECTRON TOMOGRAPHY To evaluate the effect of the
enhancement method on real experimental data with super low image contrast, we applied the method to a set of tilt series of cryo-EM images of a particle of LDL bound to CETP32. The tilt
series of cryo-ET images was acquired from a series of tilting angles from −57° to +57° at 1.5° increments, under a low-dose condition (a total dose of ~25 e−/Å2) and a magnification of 50 k
× (corresponding to 2.4 Å/pixel). The survey of cryo-ET micrographs at 3 representative angles showed the LDL and CETP particles from orthogonal views (Fig. 6A). Remarkably, the CETP’s
molecular weight was significantly below the limitations of cryo-EM; the rod-shaped CETP could scarcely be seen on the surface of LDL, and in the background, vitreous ice, such as a LDL-CETP
complex and a CETP alone particle, were masked for visualization. To confirm that the rod-shaped particles were the protein signal instead of the noise of the image, the entire tomography
(after CTF correction by the TomoCTF software package34) was aligned and reconstructed with IMOD software36. Although the IMOD 3D density maps were too noisy to distinguish the LDL and CETP
particles from the background, the projection along the Z direction of the 3D reconstruction provided sufficient contrast to evidence that the LDL alone, LDL-CETP complex, and even CETP
alone were visible (Supplemental Fig. S2). In IPET 3D reconstruction, the extremely low image contrasts (only ~0.16 to ~0.40 with an average of ~0.29) result in extremely challenging
alignments. By combining a low pass filtering at 8 Å and the enhancement method, we increased the SNRs to a range of ~0.27 to ~0.65, with an average of ~0.47. These image contrasts provided
a sufficient signal for IPET 3D reconstruction. The tilt images were gradually and iteratively aligned to their global center (Fig. 6B), and 3D density maps (after low-pass filtering at 70
Å) were reconstructed. The FSC analysis showed that the 3D resolution was ~71.0 Å based on the FSC 0.5 criterion (Fig. 6G). The 3D maps of two represented particles in the reconstructions
were displayed from two perpendicular viewing directions, wherein an ellipsoidal particle (~260.0 Å × ~220.0 Å × ~150.0 Å) with flat opposing surfaces was attached to protrusions with length
of ~110.0 Å and ~85.0 Å (Fig. 6C,D). The protrusions are slightly shorter than the crystal structure of CETP, suggesting that the portion of one distal end of CETP penetrated the surface of
LDL, which is consistent with previous observations from the negative-staining method37. This test indicated that a single molecule of 53 kDa CETP imaged by cryo-ET could be reconstructed
by using our approach. To further evaluate whether an individual CETP could be reconstructed, we repeated the above process on another local area of the tilt series, where the images
contained some free rod-shaped particles. The SNRs of the tilt series of images were only ~0.14 to ~0.28 with an average of ~0.21. After low pass filtering and enhancement, SNRs were
increased to ~0.25 to ~0.47 with an average of ~0.36. Through the IPET reconstruction process, the tilt images were gradually and iteratively aligned to their global center (Fig. 6H). The 3D
resolution was ~95.5 Å, on the basis of the 0.5 FSC criterions (Fig. 6M). The 3D reconstruction contained a large particle with rod-shaped protrusions, and some free rod-shaped particles
(Fig. 6H). The large particle was ~260.0 Å × ~260.0 Å × ~150.0 Å, and the protrusion was ~85.0 Å in length (last column in Fig. 6H). Remarkably, two rod-shaped particles were also
reconstructed (top row in Fig. 6I,J), which were ~140 to ~160 Å long that similar or slightly longer than the CETP crystal structure. The CETP molecules fit well when docking into the rod
shapes as the best fit in the density map (bottom row in Fig. 6I,J). The shape and surface of CETP were also visualized by the corresponding projections (Fig. 6K,L). The similar shapes and
dimensions of the rod-shaped particles to that of the crystal structure of CETP suggest that our enhancement method can achieve 3D reconstruction of a single molecule of 53 kDa CETP,
although the CETP molecule has a molecular weight nearly one-third the minimum limit for cryo-EM. The enhancement method was finally examined on a tilt series of DNA origami cryo-PS sample
imaged by using Gatan K2 Summit direct electron detector. This tilt series was acquired from −48° to +48° at 1.5° increments under a magnification of 19 k × (corresponding to 1.85 Å/pixel).
Three representative tilt images showed overall square-shaped DNA origamis (Fig. 7A). It should be pointed out that IPET reconstruction of these DNA origami particles was previously
published2, in which only a low-pass filter was used to enhance image contrast. Here, to evaluate our proposed enhancement algorithm, the low-pass filtering was conducted before enhancement
and IPET reconstruction. The contrasts of images were improved by both low-pass filtering and enhancement method, _i.e_., the average SNR of images increased from ~0.13 to ~0.23 by low-pass
filtering, and then increased to ~0.36 by enhancement. Using those contrast enhanced images, a 3D map at a resolution of ~97.7 Å was obtained (Fig. 7B,E). The 3D map showed an overall ~70 nm
quadrilateral shape particle (Fig. 7C) formed by four ~35-nm-long arms. By flexibly docking, an origami model could be obtained from a 3D map as a conformation of DNA origami (Fig. 7D).
Using the same protocol, another 3D map at a resolution of 97.4 Å was also obtained from another targeted particle (Fig. 7F,I). The map allowed us to determine another conformation of DNA
origami. These features of the density maps were very similar to the previous publication2, in which the low-pass filter was only used to increase the image contrast. The edge-preserving
image enhancing algorithm with the boost function was nonlinear function in changing the density of the image, which did not reflect the electron density of the protein. However, the FSC
analysis on the 2D images and 3D reconstruction did not show the influence to the resolution or structure features on both simulation and experimental results. The possibilities for this
phenomena could be the resolution obtained at current stage was not sufficiently high enough to show the influence, or the intensity obtained from EM did not truly reflect to the density of
the protein due to multiple scattering of electron beam in electron microscopic imaging38,39, and/or element electron scattering was not linearly related to the mass of element due to
elastic and inelastic scattering factors40,41,42. Nevertheless, the method increased the image contrast without reducing the 3D resolution at intermediate resolution showed the positive sign
about the method. CONCLUSION In this paper, an image contrast enhancement method was developed to increase the SNR before 3D reconstruction. The method was not a replacement method to
current filters (such as low-pass filter or band-pass filter), but can be used as a supplementary tool to support the current filters to increase the SNR by ~20%, and hence allowed for
successful 3D reconstruction of an individual particle of macromolecule. Through providing a substantial benefit for cryo-EM image reconstruction, this pre-process method may aid in the
study of protein dynamics via 3D structures determined from each individual particle for revealing the structure variety among different targeted particles of the same type of protein.
METHODS PREPARATION OF OPNS-EM, CRYO-EM, AND CRYO-PS SPECIMENS The NS specimens were prepared by the OpNS protocol3. An aliquot (∼4 μl) of the DNA-nanogold sample at a concentration of ∼20
μg ml−1 was placed on a thin carbon-coated 200-mesh copper grid (Cu-200CN, Pacific Grid-Tech, San Francisco, CA, USA) that had been glow-discharged. After ∼1 min of incubation, the excess
solution on the grid was blotted with filter paper. Then, the grid was washed with water and stained with 1% (w/v) uranyl formate on Parafilm before being dried under nitrogen. The cryo-EM
specimens were prepared as described previously32. LDL (produced by Children’s Hospital Oakland Research Institute) and CETP (produced by MERCK) were incubated at 37 °C for 15 minutes at a
molar ratio of ~4:1. We used a high concentration of CETP to ensure the combination of LDL and CETP. An aliquot (∼3 μl) of the LDL-CETP mixture was placed on a glow-discharged holey-carbon
grid (Cu-200HN, Pacific Grid-Tech, San Francisco, CA, USA). Then, the samples were flash-frozen in liquid ethane at ~90% humidity and 4 °C with a Leica EM GP rapid-plunging device (Leica,
Buffalo Grove, IL, USA) after being blotted with filter paper. The cryo-PS specimens of DNA-origami were prepared as proposed by Zhang _et al_.37. An aliquot (~4 μL) of the DNA origami
sample at a concentration of ~4 nM was placed on a glow-discharged lacey-carbon grid (LC200-Cu, Electron Microscopy Sciences, Hatfield, PA, USA) for ~1 min. The grid after washing by 1%
(w/v) uranyl formate was then flash-frozen in liquid ethane at ~90% humidity and 4 °C with the Leica rapid-plunging device. ET DATA ACQUISITION AND IMAGE PRE-PROCESSING EM imaging of
DNA-nanogold conjugates and LDL-CETP mixtures were conducted using a Zeiss Libra 120 transmission electron microscope (Carl Zeiss SMT GmbH, Oberkochen, Germany). The TEM was operated at 120
kV. A single-axis tilt series of the DNA-nanogold was collected from −60° to +60° in steps of 1.5° at a nominal magnification of 125 k × (0.94 Å/pixel). The LDL-CETP cryo-ET tilt series were
collected from −57° to +57° in steps of 1.5° at a nominal magnification of 50 k × (2.4 Å/pixel). The tilt series were acquired at ~1 μm defocus using a 4 k × 4 k Gatan UltraScan 4000 CCD
camera. For the cryo-ET, the electron dose per tilt series was within ~25 e−/Å2. Low-dose data acquisition was conducted by using the TEM tomography software (Gatan Inc., Pleasanton, CA,
USA) in Advanced Tomography mode. EM imaging of DNA origami was conducted using a FEI Tecnai TF20 TEM operated at 200 kV. A single-axis tilt series of the DNA origami was collected from −48°
to +48° in steps of 1.5° at a nominal magnification of 19 k × (1.85 Å/pixel). The tilt series were acquired at ~1 μm defocus using a Gatan K2 Summit direct electron detector, and the total
electron dose was ~50 e−/Å2. IPET 3D RECONSTRUCTION The defocus value of each tilt image was calculated by using the programs tomoctffind.exe in the TomoCTF software34. The phase of the tilt
series was corrected by the program ctfcorrect.exe in the TomoCTF software. To reconstruct the 3D structures of individual particles, we first boxed the images of the particle from each
tilt series of micrographs. The boxed images were then binned (DNA-nanogold conjugates for 3 times, LDL-CETP mixture and DNA origami for 2 times) to reduce computation time, and submitted to
IPET 3D reconstruction1. The 3D reconstructions were finally submitted to a missing-wedge correction processing. The resolution was defined on the basis of FSC, when the frequency first
decreased to a value of 0.5. The FSC curve was calculated by two 3D reconstructions generated from odd- or even-numbered index-aligned images. The SNR in both the 2D image and 3D map was
calculated using the equation SNR = (I_s_ − I_b_)/N_b_, where I_s_ is the average intensity inside the particle, I_b_ is the average intensity outside the particle, and N_b_ is the s.d. of
the noise calculated from the background (s.d. outside the particle). PROGRAM AVAILABILITY INFORMATION Software is available free of charge to academic end users. DATA AVAILABILITY The
datasets generated and/or analyses during the current study are available from the corresponding author on reasonable request. The IPET 3D density maps are available from the EM data bank
(The NS density maps of DNANG: EMD-9262 and -9263; The cryo-EM density maps of LDL-CETP: EMD-9268 and -9269; The cryo-EM density maps of CETP alone: EMD-9270 and -9271; The cryo-PS density
maps of DNA origami: EMD-9266 and -9267). Detailed experimental conditions is listed in Supplementary Table 1. REFERENCES * Zhang, L. & Ren, G. IPET and FETR: experimental approach for
studying molecular structure dynamics by cryo-electron tomography of a single-molecule structure. _PloS one_ 7, e30249, https://doi.org/10.1371/journal.pone.0030249 (2012). Article ADS CAS
PubMed PubMed Central Google Scholar * Lei, D. _et al_. Three-dimensional structural dynamics of DNA origami Bennett linkages using individual-particle electron tomography. _Nature
communications_ 9, 592, https://doi.org/10.1038/s41467-018-03018-0 (2018). Article ADS CAS PubMed PubMed Central Google Scholar * Zhang, L. _et al_. Three-dimensional structural
dynamics and fluctuations of DNA-nanogold conjugates by individual-particle electron tomography. _Nature communications_ 7, 11083, https://doi.org/10.1038/ncomms11083 (2016). Article ADS
CAS PubMed PubMed Central Google Scholar * Irobalieva, R. N. _et al_. Erratum: Structural diversity of supercoiled DNA. _Nature communications_ 6, 8851,
https://doi.org/10.1038/ncomms9851 (2015). Article CAS PubMed Google Scholar * Irobalieva, R. N. _et al_. Structural diversity of supercoiled DNA. _Nature communications_ 6, 8440,
https://doi.org/10.1038/ncomms9440 (2015). Article CAS PubMed PubMed Central Google Scholar * Zhang, X. _et al_. 3D Structural Fluctuation of IgG1 Antibody Revealed by Individual
Particle Electron Tomography. _Scientific reports_ 5, 9803, https://doi.org/10.1038/srep09803 (2015). Article PubMed PubMed Central Google Scholar * Correia, I. _et al_. The structure of
dual-variable-domain immunoglobulin molecules alone and bound to antigen. _Mabs_ 5, 364–372, https://doi.org/10.4161/mabs.24258 (2013). Article PubMed PubMed Central Google Scholar *
Jay, J. _et al_. IgG Antibody 3D Structures and Dynamics. _Antibodies_ 7, 18, https://doi.org/10.3390/antib7020018 (2018). Article Google Scholar * Zhang, M. _et al_. Assessing the
mechanisms of cholesteryl ester transfer protein inhibitors. _Biochimica et biophysica acta_ 1862, 1606–1617, https://doi.org/10.1016/j.bbalip.2017.09.004 (2017). Article CAS PubMed
Google Scholar * Yu, Y. _et al_. Polyhedral 3D structure of human plasma very low density lipoproteins by individual particle cryo-electron tomography1. _Journal of lipid research_ 57,
1879–1888, https://doi.org/10.1194/jlr.M070375 (2016). Article CAS PubMed PubMed Central Google Scholar * Lu, Z. _et al_. Molecular Architecture of Contactin-associated Protein-like 2
(CNTNAP2) and Its Interaction with Contactin 2 (CNTN2). _The Journal of biological chemistry_ 291, 24133–24147, https://doi.org/10.1074/jbc.M116.748236 (2016). Article CAS PubMed PubMed
Central Google Scholar * Zhang, M. _et al_. HDL surface lipids mediate CETP binding as revealed by electron microscopy and molecular dynamics simulation. _Scientific reports_ 5, 8741,
https://doi.org/10.1038/srep08741 (2015). Article CAS PubMed PubMed Central Google Scholar * Lu, Z. _et al_. Calsyntenin-3 molecular architecture and interaction with neurexin 1alpha.
_The Journal of biological chemistry_ 289, 34530–34542, https://doi.org/10.1074/jbc.M114.606806 (2014). Article PubMed PubMed Central Google Scholar * Zhang, L., Tong, H., Garewal, M.
& Ren, G. Optimized negative-staining electron microscopy for lipoprotein studies. _Biochimica et biophysica acta_ 1830, 2150–2159, https://doi.org/10.1016/j.bbagen.2012.09.016 (2013).
Article CAS PubMed Google Scholar * Yaroslavskij, L. P. _Digital picture processing_. _An introduction_, 9, (Springer-Verlag, 1985). * Perona, P. & Malik, J. Scale-space and edge
detection using anisotropic diffusion. _IEEE Trans. Pattern Anal. Mach. Intell._ 12, 629–639 (1990). Article Google Scholar * Aurich, V. & Weule, J. Vol. Mustererkennung. Informatik
aktuell. (eds Sagerer G., Posch S., & Kummert F.) (Springer, 1995). * Smith, S. M. & Brady, J. M. SUSAN—a new approach to low level image processing. _Int. J. Comput. Vis._ 23,
45–78, https://doi.org/10.1023/a:1007963824710 (1997). Article Google Scholar * Luo, Y., Marhoon, M., Al Dossary, S. & Alfaraj, M. Edge-preserving smoothing and applications. _The
Leading Edge_ 21, 136–158, https://doi.org/10.1190/1.1452603 (2002). Article Google Scholar * Durand, F. & Dorsey, J. Fast bilateral filtering for the display of high-dynamic-range
images. _ACM Trans. Graph._ 21, 257, https://doi.org/10.1145/566654.566574 (2002). Article Google Scholar * Li, Y., Sharan, L. & Adelson, E. H. Compressing and companding high dynamic
range images with subband architectures. _ACM Trans. Graph._ 24, 836, https://doi.org/10.1145/1073204.1073271 (2005). Article Google Scholar * Farbman, Z., Fattal, R., Lischinski, D. &
Szeliski, R. Edge-preserving decompositions for multi-scale tone and detail manipulation. _ACM Trans. Graph._ 27, 67, https://doi.org/10.1145/1399504.1360666 (2008). Article Google Scholar
* Tomasi, C. & Manduchi, R. Bilateral filtering for gray and color images. _IEEE Comput. Vis_., 839–846, https://doi.org/10.1109/iccv.1998.710815 (1998). * Lagendijk, R. L., Biemond,
J. & Boekee, D. E. Regularized iterative image restoration with ringing reduction. _IEEE Trans. Acoust._ 36, 1874–1888, https://doi.org/10.1109/29.9032 (1988). Article MATH Google
Scholar * Hertz, J., Krogh, A., Palmer, R. G. & Horner, H. Introduction to the Theory of Neural Computation. _Physics Today_ 44, 70–70, https://doi.org/10.1063/1.2810360 (1991). Article
ADS Google Scholar * Hassan, N. & Akamatsu, N. _A New Approach for Contrast Enhancement Using Sigmoid Function_. Vol. 1 (2004). * Kaspar Hollenstein, D. C. F. & Kaspar, P.
Locher. Structure of an ABC transporter in complex with its binding protein. _Nature_ 446, 213–216 (2007). Article ADS Google Scholar * Qiu, X. Y. _et al_. Crystal structure of
cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. _Nat. Struct. Mol. Biol._ 14, 106–113, https://doi.org/10.1038/nsmb1197 (2007). Article CAS PubMed
Google Scholar * Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. _J. Struct. Biol._ 128, 82–97,
https://doi.org/10.1006/jsbi.1999.4174 (1999). Article CAS PubMed Google Scholar * Frank, J. _et al_. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and
related fields. _J. Struct. Biol._ 116, 190–199 (1996). Article CAS Google Scholar * Campbell, M. G., Veesler, D., Cheng, A., Potter, C. S. & Carragher, B. 2.8 Å resolution
reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. _Elife_ 4 (2015). * Yu, Y. _et al_. Polyhedral 3D structure of human plasma very-low-density
lipoproteins by individual particle cryo-electron tomography. _J. Lipid Res._ 57, 1879–1888 (2016). Article Google Scholar * Kahlon, T. S., Shore, V. G. & Lindgren, F. T. Heterogeneity
of molecular-weight and apolipoproteins in low-density lipoproteins of healthy-human males. _Lipids_ 27, 1055–1057, https://doi.org/10.1007/Bf02535588 (1992). Article CAS PubMed Google
Scholar * Fernandez, J. J., Li, S. & Crowther, R. A. CTF determination and correction in electron cryotomography. _Ultramicroscopy_ 106, 587–596,
https://doi.org/10.1016/j.ultramic.2006.02.004 (2006). Article CAS PubMed Google Scholar * Zhang, T. _et al_. Cellular effect of high doses of silica-coated quantum dot profiled with
high throughput gene expression analysis and high content cellomics measurements. _Nano Lett._ 6, 800–808, https://doi.org/10.1021/nl0603350 (2006). Article ADS CAS PubMed PubMed Central
Google Scholar * Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. _J. Struct. Biol._ 116, 71–76,
https://doi.org/10.1006/jsbi.1996.0013 (1996). Article CAS PubMed Google Scholar * Zhang, L. _et al_. Structural basis of transfer between lipoproteins by cholesteryl ester transfer
protein. _Nat. Chem. Biol._ 8, 342–349, https://doi.org/10.1038/nchembio.796 (2012). Article CAS PubMed PubMed Central Google Scholar * Levine, Z. H. Tomography in the multiple
scattering regime of the scanning transmission electron microscope. _Applied Physics Letters_ 82, 3943–3945, https://doi.org/10.1063/1.1579116 (2003). Article ADS CAS Google Scholar *
Juffmann, T. _et al_. Multi-pass transmission electron microscopy. _Scientific reports_ 7, 1699, https://doi.org/10.1038/s41598-017-01841-x (2017). Article ADS CAS PubMed PubMed Central
Google Scholar * Colliex, C. _et al_. Electron diffraction. _C_, 259–429, https://doi.org/10.1107/97809553602060000593 (2006). Chapter Google Scholar * Peng, L. M., Ren, G., Dudarev, S.
L. & Whelan, M. J. Debye–Waller Factors and Absorptive Scattering Factors of Elemental Crystals. _Acta Crystallographica Section A Foundations of Crystallography_ 52, 456–470,
https://doi.org/10.1107/s010876739600089x (1996). Article Google Scholar * Peng, L. M., Ren, G., Dudarev, S. L. & Whelan, M. J. Robust Parameterization of Elastic and Absorptive
Electron Atomic Scattering Factors. _Acta Crystallographica Section A Foundations of Crystallography_ 52, 257–276, https://doi.org/10.1107/s0108767395014371 (1996). Article Google Scholar
Download references ACKNOWLEDGEMENTS We thank Drs Jessica M. Smith and Paul Alivisatos for DNA-nanogold sample, Dr. Lei Zhang for the negative-stained ET images, Dr. Ron Krauss for the LDL
sample, and Dr. Xiayang Qiu for the CETP sample to test these software programs. Work at the Molecular Foundry was supported by the Office of Science, Office of Basic Energy Sciences of the
U.S. Department of Energy under Contract No. DE-AC02-05CH11231. G.R. is supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (No. R01HL115153) and
the National Institute of General Medical Sciences of the National Institutes of Health (No. R01GM104427). H.W. and R.B. are supported by National Natural Science Foundation of China (No.
61601033 to H.W., 61571049 to R.B.). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * College of Information Science and Technology, Beijing Normal University, Beijing, China Hao Wu &
Rongfang Bie * The Molecular Foundry, Lawrence Berkeley National Laboratory, Berkeley, CA, 94720, USA Hao Wu, Xiaobo Zhai, Dongsheng Lei, Jianfang Liu, Yadong Yu & Gang Ren Authors * Hao
Wu View author publications You can also search for this author inPubMed Google Scholar * Xiaobo Zhai View author publications You can also search for this author inPubMed Google Scholar *
Dongsheng Lei View author publications You can also search for this author inPubMed Google Scholar * Jianfang Liu View author publications You can also search for this author inPubMed Google
Scholar * Yadong Yu View author publications You can also search for this author inPubMed Google Scholar * Rongfang Bie View author publications You can also search for this author inPubMed
Google Scholar * Gang Ren View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS This project was initiated by H.W. and G.R. H.W. programed the
software. Y.Y. collected the cryo-EM data. X.Z., D.L. and J.L. validated the program on both simulated and experimental E.T. data (including N.S. and cryo-EM images) by conducting the 3D
reconstructions and analyses. The cryo-EM sample of CETP-LDL complex was prepared and imaged by Y.Y. The results were interpreted by H.W., X.Z., D.L., J.L., R.B. and G.R. H.W. and X.Z.
drafted the initial manuscript, which was revised by D.L., J.L., R.B. and G.R. CORRESPONDING AUTHORS Correspondence to Hao Wu, Rongfang Bie or Gang Ren. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and
institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPORT INFORMATION SUPPLEMENTARY TABLE 1 RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the
article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use
is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Wu, H., Zhai, X., Lei, D. _et al._ An Algorithm for Enhancing the Image Contrast of
Electron Tomography. _Sci Rep_ 8, 16711 (2018). https://doi.org/10.1038/s41598-018-34652-9 Download citation * Received: 07 August 2017 * Accepted: 02 October 2018 * Published: 12 November
2018 * DOI: https://doi.org/10.1038/s41598-018-34652-9 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * Electron Tomography * Target Particle *
Tilt Series * Image Tilt * dB CMOS