A new phylogeny-based tribal classification of subfamily detarioideae, an early branching clade of florally diverse tropical arborescent legumes
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Detarioideae (81 genera, c. 760 species) is one of the six Leguminosae subfamilies recently reinstated by the Legume Phylogeny Working Group. This subfamily displays high
morphological variability and is one of the early branching clades in the evolution of legumes. Using previously published and newly generated sequences from four loci (_matK_-_trnK_,
_rpL16_, _trnG-trnG2G_ and ITS), we develop a new densely sampled phylogeny to assess generic relationships and tribal delimitations within Detarioideae. The ITS phylogenetic trees are
poorly resolved, but the plastid data recover several strongly supported clades, which also are supported in a concatenated plastid + ITS sequence analysis. We propose a new phylogeny-based
tribal classification for Detarioideae that includes six tribes: re-circumscribed Detarieae and Amherstieae, and the four new tribes Afzelieae, Barnebydendreae, Saraceae and Schotieae. An
identification key and descriptions for each of the tribes are also provided. SIMILAR CONTENT BEING VIEWED BY OTHERS COMPARATIVE PLASTOME GENOMICS, TAXONOMIC DELIMITATION AND EVOLUTIONARY
DIVERGENCES OF _TETRAENA HAMIENSIS_ VAR. _QATARENSIS_ AND _TETRAENA SIMPLEX_ (ZYGOPHYLLACEAE) Article Open access 08 May 2023 EXPLORING PHYLOGENETIC RELATIONSHIPS WITHIN THE SUBGENERA OF
_BAMBUSA_ BASED ON DNA BARCODES AND MORPHOLOGICAL CHARACTERISTICS Article Open access 16 May 2022 COMPARATIVE CHLOROPLAST GENOMICS AND INSIGHTS INTO THE MOLECULAR EVOLUTION OF _TANAECIUM_
(BIGNONIEAE, BIGNONIACEAE) Article Open access 01 August 2023 INTRODUCTION The Detarioideae is a monophyletic group of legumes (Leguminosae or Fabaceae) with an astonishing morphological
diversity that comprises c. 760 species in 81 genera distributed across the tropical regions of the world1,2,3,4. This lineage is one of the first branches in the legume phylogeny and it was
recently reinstated as subfamily Detarioideae Burmeist. in the new classification of the family proposed by the Legume Phylogeny Working Group3, which recognizes six subfamilies. Despite
its pantropical distribution, the majority of the detarioid generic and species diversity occurs in Africa and Madagascar (58% of genera and c. 330 spp.), followed by Central and South
America (20% of genera and c. 247 spp.), and Asia (12% of genera and c. 124 spp.)2. The Detarioideae include many ecologically important tree species in West Central African lowland
evergreen rainforests5,6,7, and in some forest types trees of this subfamily are the dominant species (e.g., _Brachystegia_ woodland, monodominant _Gilbertiodendron_ forests or
_Microberlina_ dominated groves6,8). Some Detarioideae species are also ecologically important components in lowland wet forests of the Neotropics (e.g., _Brownea_, _Copaifera_,
_Macrolobium_, and _Peltogyne_ species9,10,11). In contrast, in Asian tropical dipterocarp-dominated rainforests, although present, Detarioideae represent a modest fraction of the species
abundance and diversity12,13. Plants of this subfamily provide timber (e.g. _Aphanocalyx_, _Berlinia_, _Didelotia_, _Hymenaea, Peltogyne_ and _Tetraberlinia_), some of which are highly
valuable (e.g., species of _Guibourtia_), several species are the source of useful resins (e.g. _Copaifera_, _Hymenaea_), and _Tamarindus_ is used as a condiment for cooking5,14,15. Some
species are also part of cultural heritage, used for rituals and medicine or seen as holy trees (e.g. several species of _Brownea_16 and _Copaifera religiosa_17). Since the mid-1800, the
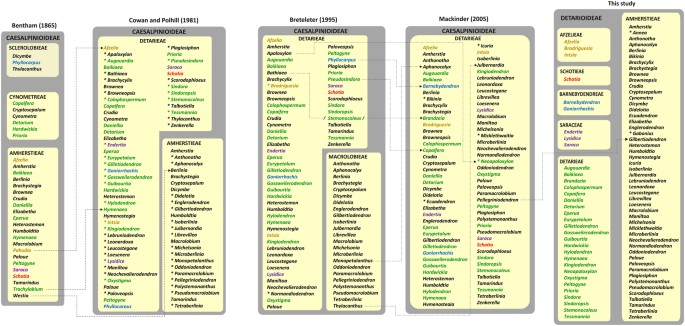
generic content of Detarioideae has remained relatively stable, but the higher level subdivision, into one or two tribes or subtribes, has fluctuated considerably (Fig. 1). Lee and
Langenheim18 provided an historical review of the tribal classification of detarioid legumes, starting with the publication of the tribe Detarieae in de Candolle’s _Prodromus_19).
Bentham20,21 established seven tribes within his 2nd legume suborder, Caesalpinieae. Two of these tribes, Amherstieae and Cynometreae, included genera ascribed to tribe Detarieae (sensu
Mackinder2). The tribe Sclerolobieae was later merged with tribe Cynometreae22,23. Based on a detailed study of seedlings of African genera, Léonard24 classified the detarioid legumes in two
tribes (Cynometreae and Amherstieae), which were later slightly modified by Heywood25 who gave priority to the name Detarieae over Cynometreae. These tribal circumscriptions were largely
followed by Cowan and Polhill26,27. Breteler28 adopted a new tribal classification for the Detarieae-Amherstieae association based on bracteole aestivation, whether valvate or imbricate, and
recognized two tribes: Detarieae (including some genera transferred from the Amherstieae) and Macrolobieae Breteler (Fig. 1). However, molecular studies subsequently showed that the
Macrolobieae is nested with genera previously recognized as part of Amherstieae29,30,31. In the _Phytochemical Dictionary of the Leguminosae_, Polhill32 accepted a single tribe Detarieae
_s.l_., and this was followed by Mackinder2 and subsequent taxonomic treatments. Phylogenetic studies have demonstrated that no previous tribal circumscriptions are supported as
monophyletic, but several well-supported clades have been resolved within Detarioideae since the first comprehensive molecular studies attempted to resolve relationships in the group29,31.
These include the Prioria, Brownea and Amherstieae clades. Subsequent studies have focused on specific clades. Wieringa and Gervais33 studied the “babijt” clade including the
_Aphanochalyx_-_Bikinia_-_Tetraberlinia_ group, which also received support from a chemical analysis34. Fougère-Danezan _et al_.35,36,37 studied the Detarieae in which they recognised the
“resin-producing Detarieae”, a group that comprises the Detarieae _s.s_. and the Prioria clade, and which produces bicyclic diterpenes36. Other phylogenetic studies have focused on subsets
of Detarioideae genera (e.g.,5,10,15,35,38,39,40,41,42). More recently Estrella _et al_.43 studied the biogeographic origin of the subfamily proposing a probable _terra firme_ African origin
in the Palaeocene with subsequent and frequent early dispersals to South America and Asia. The recently published subfamily framework for legumes3 highlighted the need for new
classifications at the supra-generic level of some of the six recognised subfamilies. Phylogeny-based classifications of taxonomically complex, ecologically diverse and morphologically
heterogeneous clades such as the Detarioideae are essential to pave the way for further taxonomic studies of genera and groups of genera, as well for tracking the course of morphological
evolution, speciation and extinction patterns, and biome shifts. The objective of the present study is to produce a new tribal classification that reflects current knowledge of phylogenetic
relationships in Detarioideae, supported by a near complete generic level sampling and a representative species level sampling. MATERIAL AND METHODS TAXON SAMPLING A total of 501 accessions,
representing 280 species of Detarioideae from 73 of the 81 genera were sampled. Additionally, two genera of subfamily Cercidoideae and one each of Duparquetioideae and Caesalpinioideae were
sampled as outgroups. This is the broadest sampling of Detarioideae species assembled to date for phylogenetic analysis (Supplementary Appendix I provides voucher information and GenBank
accession numbers). Samples collected in the field were preserved in silica gel, and other samples were obtained from dried herbarium specimens. We generated most of the sequences (including
475 sequences newly released for this study), and the sampling was completed with additional sequences produced by our research group in previous studies30,36,39,40,44 which were downloaded
from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) to complete the taxon and gene sampling. To avoid the effects of missing data no sample was included that had fewer than two loci
sequenced, and for this reason six genera that have been included in other studies (_Brachycylix_, _Lebruniodendron_, _Micklewaitia_, _Michelsonia, Neoapaloxylon, Paloveopsis_) are not
included in our analyses. We were not able to obtain material of _Leucostegane_ and _Pseudomacrolobium_ for sequencing. MOLECULAR METHODS DNA extraction of herbarium and silica gel dried
material was done using a modified protocol from Ky _et al_.45 rescaled for a total 3 mL of nucleic extraction buffer (15 mM Tris, 2 mM EDTA, 80 mm KCl, 20 mM NaCl, 2% β-mercaptoethanol,
PPVP 2%, 0.5% Trixon-X100) and the pellet was recovered in 2 ml of lysis buffer pH 8 (0.1 M Tris, 0.02 M EDTA, 1.25 M NaCl, MATAB 4%). Three plastid (_matK_-_trnK_, _rpL16_ and
_trnG_-_trnG2G_) regions and the nuclear ribosomal internal transcribed spacers (ITS/5.8 S) were amplified and sequenced. The PCR amplification mix in reaction volumes of 50 μL contained 4
units of Taq DNA polymerase, 1× Taq DNA polymerase buffer with 1.5 mmol MgCl2 (New England Biolabs, Pickering, Ontario, Canada), 200 μmol/L of each dNTP (Fermentas, Burlington, Ontario,
Canada), 3 μmol/L of each primer, and 50–100 ng of genomic DNA. For recalcitrant samples, BSA (0.1 μg/μL, New England BioLabs, Ipswich, Mass.), Tween 20 (0.03%, J-T. Baker, Phillipsburg, New
Jersey, USA), and pure DMSO (4%, Fisher Scientific, Ottawa, Ontario, Canada) were added to the mix. For samples that were difficult to amplify, we also used a nested PCR procedure described
in Gagnon _et al_.46. For the most problematic samples, including those with large mononucleotide repeats, we used a PCR protocol with Phusion Hot Start II High-Fidelity DNA polymerase
(Thermo Scientific, Waltham, Massachusetts, U.S.A.), which is more accurate and yields longer and higher-quality mononucleotide sequence reads47. For the ITS/5.8 S region, amplifications
were performed with the “AB101” and “AB102” primers48,49; conditions for the amplification follow Estrella _et al_.40. The _matK_ gene and the flanking 3′ intron region were amplified in one
fragment using the primers trnK685F and trnK2Rdet30 and the internal primers described in that study were used to sequence the most difficult samples. For _trnG-trnG2G_ and _rpL16_ we used
the primers and amplification conditions from Shaw _et al_.50, but because _rpL16_ was difficult to sequence due to a large adenine repeat, we designed a specific internal primer that we
used for sequencing (FX1: 5′-TGGATTATGAGTTGTGAAGC-3′). Sequencing was performed with Big Dye Terminator 3.1 chemistry on an ABI 3730xl DNA Analyzer (Applied Biosystems, Carlsbad, California,
USA) at the Genome Quebec facilities (Montreal, Canada). Sequences were assembled and edited with Geneious 4.8.5 (Biomatters Ltd., http://www.geneious.com). All sequences were subjected to
a Blast search51 and eliminated if they did not correspond to Leguminosae sequences in GenBank. The _matK-trnK_ matrix included 478 sequences from different accessions, the _trnG-trnG2G_
matrix included 446 sequences, the _rpL16_ included 473 sequences and the ITS/5.8 S matrix included 462 sequences. PHYLOGENETIC ANALYSES Sequence alignment was performed using MAFFT52 for
the plastid markers and SATé53,54,55 for ITS. We configured the SATé analysis following the approach described in Callahan and McPeek56 which initially estimates an alignment and tree with
MAFFT52 and FASTTREE57, decomposes the estimated tree using the longest-edge strategy into subsets no larger than 50% of the tips, aligns each subset with PRANK58, merges the PRANK
sub-alignments with MUSCLE, estimates a new tree from the merged alignment using RAxML59 under a GTRGAMMA model, and repeats this cycle of steps for 10 iterations. Finally, ambiguous sites
were removed using Gblocks60,61, allowing gap positions under stringent parameter settings. The ITS alignment from the last iteration of the SATé + Gblocks and the plastid alignments were
inspected and manually edited using Geneious 4.8.5 (Biomatters Ltd., http://www.geneious.com). The aligned _matK_-_trnK_ matrix had a total length of 1941 base pairs (bp), the
_trnG_-_trnG2G_ had a total length of 1102 bp, the _rpL_16 a total length of 1855 bp, and the _ITS_ was 1533 bp in length. Two matrices (ITS and combined plastid) were analysed separately
for exploratory purposes, and a concatenated plastid + nuclear matrix of all data containing only 7% of missing sequences was analysed using Maximum likelihood and Bayesian approaches to
generate the phylogenetic trees. Maximum likelihood analyses were carried out using RAxML v.8.0.062, on the CIPRES gateway v.3.363. The analyses were conducted using the GTRGAMMA model.
Branch support was assessed using the nonparametric bootstrap procedure, with 1000 replicates. jModelTest v.264 was used to estimate the best evolutionary model for each DNA locus
separately. Based on the Akaike information criterion, the best models identified were GTR + I + G for ITS/5.8 S, TVM + G for _matK/trnK_ and _rpL16_, and TPM1 uf + G for _trnG-trnG2G_.
Bayesian analyses were conducted in MrBayes v.3.265, but because it is not possible to specify the exact models for the three plastid regions in MrBayes, we used the reversible-jump MCMC
option, which allows sampling of different schemes of nucleotide substitution as part of the MCMC run (nst = mixed)46. The Bayesian estimation consisted of two independent runs during 50 ×
106 generations, sampling trees and parameters every 1000th generation. Each run consisted of four simultaneous Monte Carlo Markov Chains, and four swaps per generation. All sample points
prior to reaching stationarity of the chains were discarded (equivalent to discarding the first 10% generations as “burn-in”). Convergence was assessed by comparing majority rule consensus
trees from the two analyses and by using Tracer version 1.666 to compare density plots of the estimated parameters and of the likelihoods from the two analyses. RESULTS The nuclear and
combined plastid datasets converged individually in the Bayesian analyses, but the concatenated plastid + nuclear matrix did not reach convergence. The ITS analyses alone showed poor
resolution (results not shown), and although different options were tried for the ITS alignment, the sequences analysed showed signals of saturation. However, the RAxML ITS + plastid
topology generally supports the main clades recovered in the concatenated plastid analysis (Fig. S1). At the broad level, the analysis of the concatenated plastid markers resolved six major
clades. The African genus _Schotia_ is resolved as monophyletic [Fig. 2, posterior probability from the Bayesian plastid analysis (PP) = 1; Fig. S1, bootstrap support values from the RaxML
cp + ITS analysis (BS) = 100], poorly supported as sister to the American genera _Goniorrhachis_ and _Barnebydendron_ (Fig. 2). The relationship between these three genera and the
resin-producing Detarioideae is only moderately supported in the Bayesian analysis (Fig. 2, PP = 0.8). In the resin-producing Detarioideae, several strongly supported relationships are
confirmed, including the monophyly of the genus _Prioria_ sensu Breteler1, which together with _Colophospermum_ and _Hardwickia_, form a clade sister to a Daniellia clade comprised of
_Daniellia_ plus _Brandzeia_ (Fig. 2), sister to another clade formed by the Detarieae _sensu stricto_. In the Detarieae s.s. clade, most genera are supported as monophyletic, except
_Guibourtia_, _Copaifera_, and _Baikaea_, and the relationship between _Eperua_ and _Eurypetalum_ is not well resolved (Fig. 2). The Saraca and Afzelia clades appear as strongly supported
successive sister groups to the large Amherstieae clade [Figs 3 and S1, BS = 100%, PP = 1]. The Amherstieae includes most Detarioideae genera, with several moderately to well supported
clades recovered. Among these are the Brownea clade that includes seven neotropical genera (PP = 1.0, Fig. 3), a monophyletic group of three African endemic genera, _Didelotia_,
_Librevillea_ and _Gilbertiodendron_ (Fig. 4C, BS = 68%, PP = 1), and a group that includes _Microberlinia_, _Brachystegia_ and all of the “Babijt” genera (i.e. _Brachystegia_,
_Aphanocalyx_, _Bikinia_, _Icuria_, _Julbernardia_ and _Tetraberlinia_) that is only weakly supported as monophyletic (Fig. 4, S1, weak support: PP 0.61, BS < 50%). The monophyly of
several genera in the Amherstieae clade is poorly supported (e.g., _Crudia_, _Berlinia_, _Englerodendron_, _Tetraberlinia_) and a few other genera appear to be clearly polyphyletic (i.e.,
_Cynometra_). DISCUSSION THE NEW CLASSIFICATION The new Leguminosae classification proposed by the LPWG3 follows a traditional Linnaean approach, which as noted by others (e.g.,67,68,69) is
compatible and complementary to well-supported clade-based rank-free classifications (e.g. Dalbergioid clade70; inverted repeat [IR]-lacking clade,71). Because of this new subfamily level
classification, certain legume subfamilies require revised classifications. A new classification is particularly needed for the recircumscribed Caesalpinioideae that contains the
morphologically distinct mimosoid clade, and where efforts are ongoing to better resolve phylogenetic relationships and to arrive at a new taxonomic treatment3. Revising the classification
for the pantropical Detarioideae (Detarieae s.l. in Mackinder2,3) is also needed. In the past several years a number of studies have been published that aim to understand relationships and
evolution in this group (e.g.1,3,4,5,10,15,28,29,30,31,33,35,36,37,38,39,40,41,44,72,73,74,75,76) and along with the new phylogenetic analysis presented here, we are in a position to present
a formal tribal classification of Detarioideae that will provide the necessary framework to better understand the systematics and evolutionary origin of this lineage. PHYLOGENETIC EVIDENCE
Detarioideae represent an early branching lineage within Leguminosae evolution, estimated at 68–63 Ma43, and comprising six strongly supported main clades. These six clades have also been
resolved in previous studies; and here we recognize them at the tribe level: Schotieae Estrella, L.P. Queiroz & Bruneau, Barnebydendreae Estrella, L.P. Queiroz & Bruneau, Detarieae
DC., Saraceae Estrella, L.P. Queiroz & Bruneau, Afzelieae Estrella, L.P. Queiroz & Bruneau, and Amherstieae Benth. Three genera, _Schotia_, _Goniorrhachis_ and _Barnebydendron_,
always appear among the early branching clades within Detarioideae29,30,31,43, and in our analyses these are resolved as sister to the resin-producing Detarioideae, although this
relationship is weakly supported (Figs 2, 5). _Schotia_ (four species) has been consistently resolved as monophyletic in all analyses (Fig. 2;29,30,31,35,36,77) but its position within the
Detarioideae remains unresolved. Depending on the molecular marker or phylogenetic method, it appears as sister to _Goniorrhachis_ and _Barnebydendron_ (Figs 2, 5), as sister to the
resin-producing Detarioideae36 or in a polytomy at the base of the subfamily30. This unique southern African lineage is thus recognized here as the new monogeneric tribe Schotieae.
Morphologically, _Schotia_ can be differentiated from most other Detarioideae by its radially symmetrical flowers, with small bracteoles, four upright coloured sepals, five petals some of
which can be filamentous, ten mostly free stamens, and a tubular hypanthium4,78. The phylogenetic position of _Goniorrhachis_ and _Barnebydendron_, two neotropical monospecific genera, also
is not fully resolved, however the two genera consistently group together in a highly supported clade30,35,42 here recognized under the new tribe Barnebydendreae (Figs 2, 5). As noted by
Herendeen _et al_.42,79, members now allocated to the Barnebydendreae share the presence of a vein along the margin of the leaflets, a character used by Cowan and Polhill27 to discuss
subgroups within Detarieae. The two species also share a deep hypanthium4,80,81,82. Although it is possible to argue that these three genera should be included in a single tribe Schotieae,
the phylogenetic pattern obtained here and in previous studies29,30,31,43 do not allow us to unequivocally conclude that _Schotia_ forms a monophyletic group with _Goniorrhachis_ and
_Barnebydendron_. This approach with increased division at the tribal level provides a stricter phylogenetic framework for testing evolutionary hypotheses because we do not assume that the
two lineages, which morphologically are also very distinct, are necessarily sister clades. We re-circumscribed tribe Detarieae (Figs 2, 5) as equivalent to the resin-producing clade of
previous phylogenetic studies30,36,43. This clade was named subtribe Detariinae by Fougère-Danezan _et al_.35. This redefined Detarieae is now clearly circumscribed as grouping the 16 genera
of Detarieae _s.s_. (_sensu_ Fougère-Danezan _et al_.35), along with _Colophospermum_, _Hardwickia_, _Prioria_, _Daniellia_ and _Brandzeia_. As noted by Fougère-Danezan _et al_.35,36 most,
but not all, species in this clade produce a characteristic resin composed of various sesquiterpenes and diterpenes83,84. A few genera either lack resins or have never been tested for their
presence (_Sindoropsis_, _Baikaea_, _Eurypetalum_, _Stemonocoleus_, _Augouardia_, _Hardwickia_36;). Few morphological synapomorphies characterise this clade, however, the genera share a
combination of characters including: generally caducous stipules, leaves with few leaflets, bracteoles that are often caducous, ten stamens, a strong tendency to apetaly, and most
characteristically gland-dotted leaflets (the glands are also often present on the sepals). Certain generic relationships are now better supported than in previous studies. For example, the
monotypic Madagascan genus _Brandzeia_, which occurs in seasonally dry woodlands2,85, is resolved as sister to the monophyletic endemic African genus _Daniellia_ as also found by Bruneau _et
al_.30 and Fougere-Danezan _et al_.35 but with stronger support in our analyses. _Daniellia_ includes species found in both rain forest and savanna biomes86. In our analyses, a narrower
circumscription of the Prioria clade is strongly supported as monophyletic (Fig. 2). Breteler1 subsumed _Gossweilerodendron_, _Kingiodendron_ and _Oxystigma_ under a broadly defined
_Prioria_, a taxonomy that is in accordance with our analyses. Although all the previously recognized genera form monophyletic groups, some are only weakly supported lending support for a
more inclusive definition of _Prioria_ (Fig. 2), which is what we follow in our tribal classification. Despite the dense taxon sampling presented here, some intergeneric relationships remain
unclear. For example, relationships amongst _Tessmannia_, _Sindora_, _Sindoropsis_, _Detarium_ or _Copaifera_ remain unresolved (Fig. 2). Our study suggests that _Hymenaea_ may be nested
within a paraphyletic _Guibourtia_, as noted in previous studies36,43, and that together these two genera are strongly supported as sister to _Peltogyne_. Fougère-Danezan _et al_.35 noted
that the three genera have similar bifoliolate leaves with strongly asymmetrical leaflets with a primary vein close to the distal margin of the leaflet and a stipule insertion that is
lateral. The Saraca clade (Figs 3, 630;) comprises the Asian genera _Endertia_, _Lysidice_ and _Saraca_, and is here recognized as a new tribe Saraceae. These genera have in common a
tendency to occur in flooded habitats43 and together have been consistently resolved as monophyletic in previous phylogenetic studies29,30,31. _Lysidice_ and _Endertia_ share a
characteristic pollen ornamentation consisting of coarse strieae, to short anastomosing striae, to verrucate lirae87, and the three genera have bilaterally symmetrical flowers (more radially
symmetrical in _Saraca_, which lacks petals) generally with fewer than ten stamens, and staminodes often present (absent in _Endertia_)4. _Saraca_ is unusual among legumes in having an
unique floral homeotic conversion of petal primordia into stamens88. The Afzelia clade (_sensu_ Bruneau _et al_.30), recognized as the new tribe Afzelieae (Figs 3, 6), is particularly
interesting biogeographically and includes three disjunct genera. The monospecific _Brodriguesia_ is endemic to the Atlantic forests in Brazil; _Afzelia_ is a mainly African genus that is
thought to have originated in the savanna but which also includes polyploid species in forest habitats89; and _Intsia_ is found on both sides of the Indian Ocean and is likely
sea-dispersed2. _Brodriguesia_ has flowers with five almost equally sized petals whereas _Afzelia_ and _Intsia_ share a similar floral morphology with a large bilobed adaxial petal2. Despite
these divergent floral patterns, the three genera share leaves with few (and large) leaflets, each with the main vein asymmetrically displaced and a few crateriform glands near the base on
the lower surface. Tribe Amherstieae as here circumscribed was found to be monophyletic by Bruneau _et al_.30 with moderate support, and is here strongly supported as monophyletic and sister
to Afzelieae (Figs 3 and 4). The strongly supported Brownea clade (Fig. 3), has one poorly supported clade of _Brownea_ species occurring as unresolved relative to the other genera and to
the remaining Amherstieae clade lineages. The Brownea Group was initially described by Cowan and Polhill27 and considered to include 10 neotropical endemic genera. It was subsequently
redefined by Bruneau _et al_.29,31 to comprise seven genera (_Brownea_, _Browneopsis_, _Macrolobium_, _Paloue_, _Elizabetha_, _Ecuadendron_ and _Heterostemon_), with _Brachycylix_ and
_Paloveopsis_ resolved as members of the same clade by Redden _et al_.39. However, relationships among the genera of the Brownea clade remain unclear and are currently the focus of further
studies (10; R. Schley _et al_., unpublished). _Cynometra_, a pantropical genus as currently circumscribed, is well-known to be polyphyletic90 and in need of a detailed taxonomic revision
(Figs 3 and 4). Some subclades of _Cynometra_ are close relatives of the Asian genus _Maniltoa_, while another group of _Cynometra_ species are more closely related to _Hymenostegia_,
_Talbotiella_, _Loesenera_ and _Leonardoxa_. Recently two genera closely related to _Scorodophloeus_41, namely _Gabonius_ and _Annea_, were described91,92 to accommodate three species (two
sampled here) that had rendered _Hymenostegia_ polyphyletic41. As found by Estrella _et al_.40, the genus _Gilbertiodendron_, when considered to include _Pellegriniodendron_72 is supported
as monophyletic, and has been found to form a poorly supported clade with _Librevillea_ and _Didelotia_ (Fig. 4; Bruneau _et al_.30). _Anthonotha_, _Oddoniodendron_, _Isomacrolobium_ and
_Englerodendron_ have been the focus of recent taxonomic treatments73,74,93,94 but in our analyses (Fig. 4) their relationships are not clear; and only _Oddoniodendron_ is supported as
monophyletic. _Berlinia_ was monographed by Mackinder & Pennington15 who found the genus to be monophyletic in their ITS analysis and sister to a monophyletic _Isoberlinia_15. However,
generic relationships among _Berlinia_ and other Amherstieae clade genera are generally poorly resolved (Figs 4, 615,30). The “babijt” clade was described by Wieringa & Gervais33 to
group six morphologically close genera, _Brachystegia_, _Aphanocalyx_, _Bikinia_, _Icuria_, _Julbernardia_ and _Tetraberlinia_ (see also5,44), but is not supported as monophyletic in our
study (Fig. 4), because it does not include the genus _Microberlinia_, which appears as sister to _Brachystegia_ (Fig. 4; Bruneau _et al_.30). As suggested by Wieringa & Gervais33 this
clade likely also contains _Michelsonia_ and should then be called “bambijt” clade, but the latter genus could not be properly assessed in this study. The group is characterised by the
presence of 10 stamens (nine in _Aphanocalyx libellula_) and in particular bracteoles that have fully taken over the protective function of the reduced to absent sepals and that are partly
fused to the hypanthium; the pods have one or two lateral veins. Although the generic membership of Amherstieae (and the name of the clade) has varied amongst taxonomic
treatments24,27,28,30,95, there has been general consensus for recognising a cohesive group of genera based on their shared bracteole characteristics. Although the bracteoles in this clade
can be morphologically variable, in many genera they are well developed, and are larger than the sepals in bud, and thus perform the protective role normally attributed to the sepals28.
Certain Amherstieae have spectacularly showy and coloured bracteoles (Fig. 6). GAPS IN THE SAMPLING Although our study includes a broad sampling of Detarioideae taxa, eight of the 81 genera
are missing. Six of these have been sequenced for other loci in previous studies, and can be clearly assigned to the newly designated tribes. _Neoapaloxylon_ with three species endemic to
Madagascar has been sampled in the broad _matK_ LPWG phylogenetic study3 and by Fougère-Danezan _et al_.35,36 where it was found to be closely related to _Daniellia_ and _Brandzeia_ in the
newly circumscribed Detarieae. _Paloveopsis_, with a single species in Guyana and Brazil, and the monospecific _Brachycylix_ endemic to Colombia, were included in the study by Redden _et
al_. (39; R. Schley _et al_., unpublished), and found to be closely related to _Paloue_ and _Ecuadendron_, respectively, both in the Brownea clade of Amherstieae. _Lebruniodendron_ with a
single species endemic to West Central Africa was resolved as sister to _Crudia_ and _Neochevalierodendron_41 as is best considered part of Amherstieae, as is _Micklethwaitia_2,96, a
monospecific genus endemic to Mozambique and previously treated under _Cynometra_, which was found to be closely related to _Gabonius_2,96. The monospecific _Michelsonia_ from Congo
(Kinshasa) was found to belong to the “babijt” clade (_sensu_33) within Amherstieae based on a single plastid _psbA-trnH_ sequence44 confirming the morphological analysis by Wieringa5, but
the exact relationship of this poorly sampled species remains unresolved. Two genera have never been sequenced because of a lack of material. Nevertheless, _Pseudomacrolobium_, which
includes a single species from Congo (Kinshasa), was considered by Mackinder2 to be part of Amherstieae, and _Leucostegane_ (2 spp. from Malesia), is considered to be closely related to
_Saraca_ and _Lysidice_2 and can confidently be assigned to Saraceae, based on morphological characters. SYSTEMATIC TREATMENT Subfamily Detarioideae Burmeist., Handb. Naturgesch.: 319. 1837,
emend. LPWG, Taxon 66 (1): 44–77. 2017. Currently 81 genera and c. 760 species1,2,3,43, almost exclusively tropical with genera present in Central and South America, Africa and South East
Asia; and the genus _Schotia_ in sub-tropical South Africa. KEY TO DETARIOIDEAE TRIBES 1. Leaflets generally with translucent gland dots; cut bark exudes resin………DETARIEAE 1. Leaflets
lacking translucent gland dots; cut bark generally not exuding resin………2 2. Bracteoles well-developed (usually persistent), often enveloping the calyx in bud………AMHERSTIEAE 2. Bracteoles
well-developed or not, generally caduceus………3 3. Functional stamens generally fewer than 10, staminodes often present………SARACEAE 3. Functional stamens generally 10, staminodes absent………4 4.
Flower hypanthium shortly tubular, stipe free………BARNEBYDENDREAE 4. Flower hypanthium shallow, stipe adnate to hypanthium………5 5. Flowers radially symmetrical………SCHOTIEAE 5. Flowers
bilaterally symmetrical………AFZELIEAE - Tribe SCHOTIEAE Estrella, L.P. Queiroz & Bruneau, TRIBUS NOV. Type: _Schotia_ Jacq. Included genera (1): _Schotia_ Jacq. (4 species) (Fig. 5a,b).
Leaflets alternate or opposite, petiolulate, sometimes sessile, lacking translucent gland dots. Flowers radially symmetrical; bracteoles small, caducous, not protecting the bud; sepals 4 (5
initiated but the two adaxial fused at maturity88), well developed; petals generally 5, but 1 or more may be reduced or narrow; stamens 10, free or joined at the base; stipe short, adnate to
hypanthium. Fruits dehiscent, but the sutural frame persistent. Seeds arillate. Distribution: tropical and subtropical South Africa, generally in the drier succulent biome14. - Tribe
BARNEBYDENDREAE Estrella, L.P. Queiroz & Bruneau, TRIBUS NOV. Type: _Barnebydendron_ J.H.Kirkbr. Included genera (2): _Barnebydendron_ J.H. Kirkbr. (1), _Goniorrhachis_ Taub. (1) (Fig.
5c,d). Leaflets opposite, petiolulate, lacking translucent gland dots. Flowers weakly (Goniorrhachis) or strongly (Barnebydendron) bilaterally symmetrical; bracteoles well developed but not
showy, caducous to briefly persistent, not protecting the bud; sepals 4, well developed; petals (3-)5, subequal to 2–3 well developed and the remaining petals reduced; stamens 10, free in
two whorls (Goniorrhachis) or diadelphus (9 + 1) (Barnebydendron), bent in bud becoming upcurved at anthesis; stipe free in a shortly tubular hypanthium. Fruits indehiscent, samaroid, with a
rib on each side parallel to the upper margin. Seeds exarillate. Distribution: from Central America (Guatemala to Panama) to South America (Bolivia to the Atlantic coast of Brazil). The two
species are found in seasonally dry tropical forest, in the succulent biome14. - Tribe DETARIEAE DC., Prodr. 2: 521. 1825. Type: _Detarium_ Juss. Included genera (21): _Augouardia_ Pellegr.
(1), _Baikiaea_ Benth. (4), _Brandzeia_ Baill. (1), _Colophospermum_ J. Kirk ex J. Léonard (1), _Copaifera_ L. (c. 35), _Daniellia_ Benn. (10), _Detarium_ Juss. (3), _Eperua_ Aubl. (14),
_Eurypetalum_ Harms (2), _Gilletiodendron_ Vermoesen (5), _Guibourtia_ Benn. (14), _Hardwickia_ Roxb. (1), _Hylodendron_ Taub. (1), _Hymenaea_ L. (14), _Neoapaloxylon_ Rauschert (3),
_Peltogyne_ Vogel (c. 25), _Prioria_ Griseb. (including _Gossweilerodendron_ Harms, _Kingiodendron_ Harms and _Oxystigma_ Harms, c. 14 species), _Sindora_ Miq. (c. 20), _Sindoropsis_ J.
Léonard (1), _Stemonocoleus_ Harms (1) and _Tessmannia_ Harms (c. 12) (Fig. 5e–h). Leaflets opposite to alternate, petiolulate, often with translucent gland dots, species characterized by
the ability to produce bicyclic diterpenes. Flowers with a weak bilateral symmetry; bracteoles small, caducous, not protecting the bud; sepals 4–5 per flower, well developed; petals 0–5,
usually equal; stamens generally 10, but sometimes reduced to 3–4 (_Augouardia_ and _Stemonocoleus_) or up to 25 (_Colophospermum_), usually several of them partially joined for variable
lengths; stipe absent or adnate to hypanthium. Fruits dehiscent or indehiscent. Seeds arillate or exarillate. Distribution: pantropical, but 11 genera restricted to continental Africa, two
restricted to Madagascar, two to Asia and two to the neotropics. Broadly distributed, genera in this tribe tend to occur in wet tropical evergreen forests14. - Tribe SARACEAE Estrella, L.P.
Queiroz & Bruneau, TRIBUS NOV. Type: _Saraca_ L. Included genera (4): _Endertia_ Steenis & de Wit (1), _Leucostegane_ Prain (2), _Lysidice_ Hance (2), _Saraca_ L. (c. 11) (Fig.
6a,b). Leaflets opposite or subopposite, petiolulate to sessile, lacking translucent gland dots. Flowers bilaterally symmetrical (radially symmetrical in _Saraca_); bracteoles small to large
and showy, usually not protecting the bud; pedicels articulated; sepals 4, well developed, imbricate; petals 0–5, variable in size and shape, generally with 1–3 well developed, remaining
vestigial or absent; stamens 2 [3–8(−10) in _Saraca_], free, usually 3–8 staminodes also present; ovary stipe free to adnate to the hypanthium wall. Fruits dehiscent with twisting valves.
Seeds exarillate. Distribution: from Indo-China to Malesia, extending to the Pacific islands, generally in lowland tropical forest, within the rainforest biome14. - Tribe AFZELIEAE Estrella,
L.P. Queiroz & Bruneau, TRIBUS NOV. Type: _Afzelia_ Sm. Included genera (3): _Afzelia_ Sm. (c. 11), _Brodriguesia_ R.S. Cowan (1), _Intsia_ Thouars (3) (Fig. 6c,d). Leaflets opposite,
petiolulate, lacking translucent gland dots. Flowers bilaterally symmetrical; bracteoles well developed, caducous, not protecting the flower; sepals 4, well developed, imbricate, only 2
visible in bud; petals 5, one large petal and 4 reduced (Afzelia and Intsia) or 5 well developed (Brodriguesia); stamens 3 (Intsia), 7(−9) (Afzelia) or 10 (Brodriguesia), free or basally
connate; stipe adnate to hypanthium. Fruits dehiscent but valves not becoming twisted. Seeds with a cupular or annular aril, or aril-like structure. Distribution: pantropical. _Intsia_ and
_Brodriguesia_ are distributed within the rainforest biome, meanwhile _Afzelia_ species appear within rainforest and the grassland biomes14,89. - Tribe AMHERSTIEAE Benth., J. Bot. (Hooker)
2: 73. 1840. Type: Amherstia Wall. Included genera (50): _Amherstia_ Wall. (1), _Annea_ Mackinder & Wieringa (2), _Anthonotha_ P. Beauv. (c. 30), _Aphanocalyx_ Oliver (14), _Berlinia_
Sol. ex Hook. f. (c. 17), _Bikinia_ Wieringa (10), _Brachycylix_ (Harms) R.S. Cowan (1), _Brachystegia_ Benth. (c. 26), _Brownea_ Jacq. (c. 12), _Browneopsis_ Huber (6), _Crudia_ Schreb. (c.
55), _Cryptosepalum_ Benth. (c. 11), _Cynometra_ L. (c. 90), _Dicymbe_ Spruce ex Benth. & Hook. f. (c. 20), _Didelotia_ Baill. (c. 12), _Ecuadendron_ D.A. Neill (1), _Elizabetha_
Schomb. ex Benth. (c. 11), _Englerodendron_ Harms (1), _Gabonius_ Wieringa & Mackinder (1), _Gilbertiodendron_ J. Léonard (c. 30), _Heterostemon_ Desf. (7), _Humboldtia_ Vahl (6),
_Hymenostegia_ (Benth.) Harms (c. 16), _Icuria_ Wieringa (1), _Isoberlinia_ Craib & Stapf ex Holland (c. 5), _Isomacrolobium_ Aubrév. & & Pellegr. (12), _Julbernardia_ Pellegr.
(c. 11), _Lebruniodendron_ J. Léonard (1), _Leonardoxa_ Aubrév. (1), _Librevillea_ Hoyle (1), _Loesenera_ Harms (4), _Macrolobium_ Schreb. (c. 80), _Maniltoa_ Scheff. (c. 25), _Michelsonia_
Hauman (1), _Micklethwaitia_ G.P. Lewis & Schrire (1), _Microberlinia_ A. Chev. (2), _Neochevalierodendron_ J. Léonard (1), _Normandiodendron_ J. Léonard (2), _Oddoniodendron_ De Wild.
(c. 3), _Paloue_ Aubl. (4), _Paloveopsis_ R.S. Cowan (1), _Paramacrolobium_ J.Léonard (1), _Plagiosiphon_ Harms (5), _Polystemonanthus_ Harms (1), _Pseudomacrolobium_ Hauman (1),
_Scorodophloeus_ Harms (3), _Talbotiella_ Baker f. (8), _Tamarindus_ L. (1), _Tetraberlinia_ (Harms) Hauman (7) and _Zenkerella_ Taub. (c. 5) (Fig. 6e–h). Leaflets opposite or alternate,
petiolulate to sessile, lacking translucent gland dots. Flowers bilaterally to radially symmetrical; bracteoles variable, but often well developed, and becoming larger than the sepals/calyx
in flower bud; sepals (0-) 4–5 (−10), occasionally in some genera the two adaxial ones (partly) joined; petals variable, (0-) 5 (−6) often one or two petals enlarged, the remaining ones
reduced or absent; stamens extremely variable, generally 3–10 but up to 80 (e.g., in _Maniltoa_), free or basally connate, often diadelphus, sometimes staminodia also present; stipe of the
ovary free or adnate to hypanthium wall. Fruits mostly explosively dehiscent, or indehiscent (_Tamarindus_). Seeds exarillate. Distribution: predominantly pantropical, but with 34 genera
restricted to continental Africa and nine to Central and South America. genera in this tribe tend to occur in wet tropical evergreen forests14. DATA AVAILABILITY The sequences used in this
study are available for download from the GenBank database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/genbank/). See Supplementary Appendix I for the
accession numbers of all samples included. REFERENCES * Breteler, F. J. A revision of _Priori_a, including _Gossweilerodendro_n, _Kingiodendro_n, _Oxystigm_a, and _Pterygopodiu_m
(Leguminosae-Caesalpinioideae-Detarieae) with emphasis onAfrica. _Wageningen Agr. Univ. Pap._ 99, 1–61 (1999). Google Scholar * Mackinder, B. In _Legumes of the World_ (eds Lewis, G. P.,
Schrire, B., MacKinder, B. & Lock, M.) Detarieae _sensu lato_. 69–109 (Royal Botanic Gardens, 2005). * LPWG. A new subfamily classification of the Leguminosae based on a taxonomically
comprehensive phylogeny. _Taxon_ 66, 44–77 (2017). * Bruneau, A., Klitgaard, B. B., Prenner, G., Fougère-Danezan, M. & Tucker, S. C. Floral evolution in the Detarieae (Leguminosae):
phylogenetic evidence for labile floral development in an early-diverging legume lineage. _Int. J. Plant Sci._ 175, 392–417 (2014). Article Google Scholar * Wieringa, J. J.
_Monopetalanthus_ exit: a systematic study of _Aphanocalyx_, _Bikinia_, _Icuria_, _Michelsonia_ and _Tetraberlinia_ (Leguminosae, Caesalpinioideae). _Wageningen Agr. Univ. Pap._ 99-4, 1–320
(1999). Google Scholar * Newbery, D. M., Van Der Burgt, X. M., Worbes, M. & Chuyong, G. B. Transient dominance in a central African rain forest. _Ecol. Monogr._ 83, 339–382 (2013).
Article Google Scholar * Burkill, H. M. _The useful plants of West Tropical Africa: Families J - L_ Vol. 3 (Royal Botanic Gardens, 1995). * White, F. _The vegetation of Africa: a
descriptive memoir to accompany the Unesco/AETFAT/UNSO vegetation map of Africa_. (Unesco, 1983). * Cowan, R. S. A taxonomic revision of the genus _Macrolobium_
(Leguminosae-Caesalpinioideae). _Mem. N. Y. Bot. Gard. v._ 8, 257–342 (1953). Google Scholar * Murphy, B., de la Estrella, M., Schley, R., Forest, F. & Klitgaard, B. On the monophyly of
_Macrolobium_ Schreb., an ecologically diverse neotropical tree genus (Fabaceae-Detarioideae). _Int. J. Plant Sci_. (2018). * ter Steege, H. _et al_. Hyperdominance in the Amazonian tree
flora. _Science_ 342, 1243092, https://doi.org/10.1126/science.1243092 (2013). Article PubMed Google Scholar * Ghazoul, J. _Dipterocarp biology, ecology, and conservation_. (Oxford
University Press, 2016). * LaFrankie, J. V. _Trees of tropical Asia: an illustrated guide to diversity_. (Black Tree Publications, 2010). * Lewis, G., Schrire, B., Mackinder, B. & Lock,
M. _Legumes of the World_. (Royal Botanic Gardens, 2005). * Mackinder, B. & Pennington, R. T. Monograph of _Berlinia_ (Leguminosae). _Syst. Bot. Monogr._ 91, 1–117 (2011). Google Scholar
* Klitgaard, B. B. Ecuadorian _Brownea a_nd _Browneopsis_ (Leguminosae-Caesalpinioideae): taxonomy, palynology, and morphology. _Nordic J. Bot._ 11, 433–449 (1991). Article Google Scholar
* Quiroz, D. & van Andel, T. Evidence of a link between taboos and sacrifices and resource scarcity of ritual plants. _J. Ethnobiol. Ethnomed._ 11, 5,
https://doi.org/10.1186/1746-4269-11-5 (2015). Article PubMed PubMed Central Google Scholar * Lee, Y. T. & Langenheim, J. H. Systematics of the genus _Hymenaea_ L. (Leguminosae,
Caesalpinioideae, Detarieae). _University of California Publications in Botany_ 69, 1–109 (1975). Google Scholar * de Candolle, A. P. _Prodromus systematis naturalis regni vegetabilis,
sive, Enumeratio contracta ordinum generum specierumque plantarum huc usque cognitarium, juxta methodi naturalis, normas digesta auctore Aug. Pyramo de Candolle_. Vol. 2 (Sumptibus Sociorum
Treuttel et Würtz, 1825). * Bentham, G. Contributions towards a Flora of South America — Enumeration of plants collected by Mr Schomburgk in British Guiana. _J. Bot._ 2, 127–146 (1840).
Google Scholar * Bentham, G. In _Genera Plantarum_ Vol. 1 (2) (eds Bentham, G. & Hooker, J. D.) 434–600 (L. Reeve & co., 1865). * Baker, E. G. _The Leguminosae of tropical Africa_
(Erasmus Press, 1926). * Dwyer, J. D. Rapport entre stipe et coupe réceptaculaire dans la classification des Amherstieae. _Proceedings of the VIII International Botanical Congress_ 2–6,
51–54 (1954). Google Scholar * Léonard, J. Genera des Cynometreae et des Amherstieae africaines (Leguminosae - Caesalpinioideae): essai de blastogenie appliquee a la systematique. _Mém. Cl.
Sci. Acad. Roy. Sci. Belgique, (8vo)._ 30, 1–314 (1957). Google Scholar * Heywood, V. H. In _Chemotaxonomy of the_ Leguminosae (eds Harborne, J. B., Boulter, D. & Turner, B. L.) 1–29
(Academic Press, 1971). * Cowan, R. S. & Polhill, R. In _Advances in Legume Systematics_ Vol. 1 (eds Polhill, R. & Raven, P. H.) Tribe 4. Detarieae DC. (1825). 117–134 (Royal Botanic
Gardens, 1981). * Cowan, R. S. & Polhill, R. In _Advances in Legume Systematics_ Vol. 1 (eds Polhill, R. & Raven, P. H.) Tribe 5. Amherstieae Benth emend. J. Léonard (1957). 135–142
(Royal Botanic Gardens, 1981). * Breteler, F. J. In _Advances in legume systematics_ Vol. 7 (eds Crisp, M.D. & Doyle, J. J.) The boundary between Amherstieae and Detarieae
(Caesalpinioideae). 53–61 (Royal Botanic Gardens, 1995). * Bruneau, A., Forest, F., Herendeen, P. S., Klitgaard, B. B. & Lewis, G. P. Phylogenetic relationships in the Caesalpinioideae
(Leguminosae) as inferred from chloroplast _trnL_ intron sequences. _Syst. Bot._ 26, 487–514 (2001). Google Scholar * Bruneau, A., Mercure, M., Lewis, G. P. & Herendeen, P. S.
Phylogenetic patterns and diversification in the caesalpinioid legumes. _Botany_ 86, 697–718 (2008). Article CAS Google Scholar * Bruneau, A., Breteler, F. J., Wieringa, J. J., Gervais,
G. Y. F. & Forest, F. In _Advances in Legume Systematics_ Vol. 9 (eds Herendeen, P. S. & Bruneau, A.) Phylogenetic relationships in tribes Macrolobieae and Detarieae as inferred from
chloroplast trnL intron sequences. 121–149 (Royal Botanic Gardens, 2000). * Polhill, R. In _Phytochemical dictionary of the Leguminosae. Plants and their constituents_ Vol. 1 (eds Bisby, F.
A., Buckingham, J. & Harborne, J. B.) Classification of the Leguminosae. xxxv-xlvii (Chapman & Hall, 1994). * Wieringa, J. J. & Gervais, G. Y. F. In _Advances in Legume
Systematics, Part_ 10_, Higher_ Level _Systematics_ (eds Klitgaard, B. & Bruneau, A.) Phylogenetic analyses of combined morphological and molecular data sets on the
Aphano-calyx-Bikinia-Tetraberlinia group (Leguminosae, Caesalpinioideae, Detarieae s. l.). 181–196 (Royal Botanic Gardens, 2003). * Kite, G. C. & Wieringa, J. J. Hydroxypipecolic acids
and hydroxyprolines as chemical characters in _Aphanocalyx_, _Bikinia_ and _Tetraberlinia_ (Leguminosae: Caesalpinioideae): support for the segregation of Monopetalanthus. _Biochem. Syst.
Ecol._ 31, 279–292 (2003). Article CAS Google Scholar * Fougère-Danezan, M., Herendeen, P. S., Maumont, S. & Bruneau, A. Morphological evolution in the variable resin-producing
Detarieae (Fabaceae): Do morphological characters retain a phylogenetic signal? _Ann. Bot._ 105, 311–325 (2010). Article PubMed Google Scholar * Fougère-Danezan, M., Maumont, S. &
Bruneau, A. Relationships among resin-producing Detarieae s.l. (Leguminosae) as inferred by molecular data. _Syst. Bot._ 32, 748–761 (2007). Article Google Scholar * Fougère-Danezan, M.,
Maumont, S. & Bruneau, A. In _Advances in legume systematics_, Part _10_. (eds Klitgaard, B. & Bruneau, A.) Phylogenetic relationships in resin-producing Detarieae inferred from
molecular data and preliminary results for a biogeographic hypothesis. 161–180 (Royal Botanic Gardens, 2003). * Redden, K. M. & Herendeen, P. S. Morphology and phylogenetic analysis of
_Paloue_ and related genera in the Brownea clade (Detarieae, Caesalpinioideae). _Int. J. Plant Sci._ 167, 1229–1246 (2006). Article Google Scholar * Redden, K. M., Herendeen, P. S.,
Wurdack, K. J. & Bruneau, A. Phylogenetic relationships of the northeastern South American Brownea clade of tribe Detarieae (Leguminosae: Caesalpinioideae) based on morphology and
molecular data. _Syst. Bot._ 35, 524–533 (2010). Article Google Scholar * de la Estrella, M. _et al_. Phylogenetic analysis of the African genus _Gilbertiodendron_ J. Léonard and related
genera (Leguminosae-Caesalpinioideae-Detarieae). _Int. J. Plant Sci._ 175, 975–985 (2014). Article Google Scholar * Mackinder, B. A. _et al_. The tropical African legume _Scorodophloeus_
clade includes two undescribed _Hymenostegia_ segregate genera and _Micklethwaitia_, a rare, monospecific genus from Mozambique. _S. Afr. J. Bot._ 89, 156–163 (2013). Article Google Scholar
* Herendeen, P. S., Bruneau, A. & Lewis, G. In _Advances in legume systematics: part 10. Higher level systematics_ (eds Klitgaard, B. & Bruneau, A.) Phylogenetic relationships in
caesalpinioid legumes: a preliminary analysis based on morphological and molecular data. 37–62 (Royal Botanic Gardens, 2003). * de la Estrella, M., Forest, F., Wieringa, J. J.,
Fougère-Danezan, M. & Bruneau, A. Insights on the evolutionary origin of Detarioideae, a clade of ecologically dominant tropical African trees. _New Phytol._ 214, 1722–1735,
https://doi.org/10.1111/nph.14523 (2017). Article PubMed Google Scholar * Gervais, G. Y. F. & Bruneau, A. Phylogenetic analysis of a polyphyletic African genus of Caesalpinioideae
(Leguminosae): _Monopetalanthus_ Harms. _Plant Syst. Evol._ 235, 19–34 (2002). Article Google Scholar * Ky, C. L. _et al_. Interspecific genetic linkage map, segregation distortion and
genetic conversion in coffee (_Coffea sp_.). _Theor. Appl. Genet._ 101, 669–676 (2000). Article CAS Google Scholar * Gagnon, E., Hughes, C. E., Lewis, G. P. & Bruneau, A. A new
cryptic species in a new cryptic genus in the _Caesalpinia_ group (Leguminosae) from the seasonally dry inter-Andean valleys of South America. _Taxon_ 64, 468–490 (2015). Article Google
Scholar * Fazekas, A. J., Steeves, R. & Newmaster, S. G. Improving sequencing quality from PCR products containing long mononucleotide repeats. _Biotechniques_ 48, 277–281 (2010).
Article CAS PubMed Google Scholar * Sun, Y., Skinner, D. Z., Liang, G. H. & Hulbert, S. H. Phylogenetic analysis of _Sorghum_ and related taxa using internal transcribed spacers of
Nuclear Ribosomal DNA. _Theor. Appl. Genet._ 89, 26–32 (1994). Article CAS PubMed Google Scholar * Douzery, E. J. P. _et al_. Molecular phylogenetics of _Diseae_ (Orchidaceae): A
contribution from nuclear ribosomal ITS sequences. _Am. J. Bot._ 86, 887–899 (1999). Article CAS PubMed Google Scholar * Shaw, J. _et al_. The tortoise and the hare II: Relative utility
of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. _Am. J. Bot._ 92, 142–166 (2005). Article CAS PubMed Google Scholar * Altschul, S. F., Gish, W., Miller, W., Myers,
E. W. & Lipman, D. J. Basic local alignment search tool. _J. Mol. Biol._ 215, 403–410 (1990). Article CAS PubMed Google Scholar * Katoh, K. & Standley, D. M. MAFFT Multiple
sequence alignment software version 7: improvements in performance and usability. _Mol. Biol. Evol._ 30, 772–780 (2013). Article CAS PubMed PubMed Central Google Scholar * Liu, K.,
Raghavan, S., Nelesen, S., Linder, C. R. & Warnow, T. Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. _Science_ 324 (2009). * Yu, J., Holder,
M.T., Sukumaran, J., Mirarab, S. & Oaks, J. SATé version v. 2.2.7 http://phylo.bio.ku.edu/software/sate/sate.html (2013). * Liu, K. _et al_. SATe-II: Very fast and accurate simultaneous
estimation of multiple sequence alignments and phylogenetic trees. _Syst. Biol._ 61, 90–106 (2012). Article PubMed Google Scholar * Callahan, M. S. & McPeek, M. A. Multi-locus
phylogeny and divergence time estimates of _Enallagma_ damselflies (Odonata: Coenagrionidae). _Mol. Phylogenet. Evol._ 94, 182–195 (2016). Article PubMed Google Scholar * Price, M. N.,
Dehal, P. S. & Arkin, A. P. FastTree 2-Approximately maximum-likelihood trees for large alignments. _Plos One_ 5, e9490, https://doi.org/10.1371/journal.pone.0009490 (2010). Article ADS
PubMed PubMed Central Google Scholar * Loytynoja, A. & Goldman, N. An algorithm for progressive multiple alignment of sequences with insertions. _Proc. Natl. Acad. Sci. USA_ 102,
10557–10562 (2005). Article ADS PubMed PubMed Central Google Scholar * Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed
models. _Bioinformatics_ 22, 2688–2690 (2006). Article CAS PubMed Google Scholar * Talavera, G. & Castresana, J. Improvement of phylogenies after removing divergent and ambiguously
aligned blocks from protein sequence alignments. _Syst. Biol._ 56, 564–577 (2007). Article CAS PubMed Google Scholar * Castresana, J. Selection of conserved blocks from multiple
alignments for their use in phylogenetic analysis. _Mol. Biol. Evol._ 17, 540–552 (2000). Article CAS PubMed Google Scholar * Stamatakis, A. RAxML version 8: a tool for phylogenetic
analysis and post-analysis of large phylogenies. _Bioinformatics_ 30, 1312–1313, https://doi.org/10.1093/bioinformatics/btu033 (2014). Article CAS PubMed PubMed Central Google Scholar *
Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In _Proceedings of the Gateway Computing Environments Workshop
(GCE)_. 1–8 (New Orleans: IEEE. 2010). * Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. _Nat. Methods_ 9, 772–772
(2012). Article CAS PubMed PubMed Central Google Scholar * Ronquist, F. _et al_. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space.
_Syst. Biol._ 61, 539–542 (2012). Article PubMed PubMed Central Google Scholar * Rambaut, A., Suchard, M. A., Xie, D. & Drummond, A. J. Tracer, version 1.6. Available from,
http://tree.bio.ed.ac.uk/software/tracer/, (Accessed: 1st February 2017) (2014). * Wojciechowski, M. F. Towards a new classification of Leguminosae: naming clades using non-Linnaean
phylogenetic nomenclature. _S. Afr. J. Bot._ 89, 85–93 (2013). Article Google Scholar * Cantino, P. D. _et al_. Towards a phylogenetic nomenclature of Tracheophyta. _Taxon_ 56, 822–846
(2007). Article Google Scholar * Li, B. _et al_. A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. _Sci. Rep._ 6, 34343,
https://doi.org/10.1038/srep34343 (2016). Article ADS CAS PubMed PubMed Central Google Scholar * Lavin, M. _et al_. The dalbergioid legumes (Fabaceae): delimitation of a pantropical
monophyletic clade. _Am. J. Bot._ 88, 503–533 (2001). Article CAS PubMed Google Scholar * Wojciechowski, M. F., Lavin, M. & Sanderson, M. J. A phylogeny of legumes (Leguminosae)
based on analysis of the plastid _matK_ gene resolves many well-supported subclades within the family. _Am. J. Bot._ 91, 1846–1862 (2004). Article CAS PubMed Google Scholar * de la
Estrella, M., Devesa, J. A. & Wieringa, J. J. A morphological re-evaluation of the taxonomic status of the genus _Pellegriniodendron_ (Harms) J. Leonard
(Leguminosae-Caesalpinioideae-Detarieae) and its inclusion in _Gilbertiodendron_ J. Leonard. _S. Afr. J. Bot._ 78, 257–265 (2012). Article Google Scholar * Breteler, F. J. Revision of the
African genus Isomacrolobium (Leguminosae, Caesalpinioideae). _Plant Ecol. Evol_ 144, 64–81 (2011). Article Google Scholar * Breteler, F. J. Revision of the African genus _Anthonotha_
(Leguminosae, Caesalpinioideae). _Plant Ecol. Evol_, 70–99 (2010). * LPWG. Legume phylogeny and classification in the 21st century: Progress, prospects and lessons for other species-rich
clades. _Taxon_ 62, 217–248 (2013). * LPWG. Towards a new classification system for legumes: Progress report from the 6th International Legume Conference. _S. Afr. J. Bot_. 89, 3–9 (2013). *
Ramdhani, S., Cowling, R. M. & Barker, N. P. Phylogeography of _Schotia_ (Fabaceae): recent evolutionary processes in an ancient thicket biome lineage. _Int. J. Plant Sci._ 171, 626–640
(2010). Article Google Scholar * Tucker, S. C. Floral development in _Schotia_ and _Cynometra_ (Leguminosae: Caesalpinioideae: Detarieae). _Am. J. Bot._ 88, 1164–1180 (2001). Article CAS
PubMed Google Scholar * Herendeen, P. S., Lewis, G. P. & Bruneau, A. Floral morphology in Caesalpinioid legumes: Testing the monophyly of the “Umtiza clade”. _Int. J. Plant Sci._
164, S393–S407 (2003). Article Google Scholar * Warwick, M. C. & Lewis, G. P. & Lima, H. C. d. A reappraisal of _Barnebydendron_ (Leguminosae: Caesalpinioideae: Detarieae). _Kew
Bull._ 63, 143–149 (2008). Article Google Scholar * Prenner, G. & Cardoso, D. Flower development of _Goniorrhachis marginata_ reveals new insights into the evolution of the florally
diverse detarioid legumes. _Ann. Bot._ 119, 417–432 (2017). Article PubMed Google Scholar * Tucker, S. C. Comparative floral ontogeny in Detarieae (Leguminosae: Caesalpinioideae). 2.
Zygomorphic taxa with petal and stamen suppression. _Am. J. Bot._ 89, 888–907 (2002). Article PubMed Google Scholar * Langenheim, J. H. In _Advances in legume systematics_ Vol. 2 (eds
Polhill, R. M. & Raven, P. H.) Terpenoids in the Leguminosae. 627–656 (Royal Botanic Gardens, 1981). * Langenheim, J. H. _Plant resins: chemistry, evolution, ecology, and ethnobotany_.
(Timber Press, 2003). * Du Puy, D. J. _et al_. _The Leguminosae of Madagascar_ (Royal Botanic Gardens 2002). * de la Estrella, M., Aedo, C., Mackinder, B. & Velayos, M. Taxonomic
revision of _Daniellia_ (Leguminosae: Caesalpinioideae). _Syst. Bot._ 35, 296–324 (2010). Article Google Scholar * Banks, H. & Klitgaard, B. In _Advances in legume systematics, part 9_
(eds. Herendeen, P. S. & Bruneau, A.) Palynological contribution to the systematics of detarioid legumes. 79–106 (Royal Botanic Gardens, 2000). * Tucker, S. C. Floral development and
homeosis in _Saraca_ (Leguminosae: Caesalpinioideae: Detarieae). _Int. J. Plant Sci._ 161, 537–549 (2000). Article Google Scholar * Donkpegan, A. S. L. _et al_. Evolution in African
tropical trees displaying ploidy-habitat association: the genus _Afzelia_ (Leguminosae). _Mol. Phylogenet. Evol._ 107, 270–281, https://doi.org/10.1016/j.ympev.2016.11.004 (2017). Article
PubMed Google Scholar * Radosavljevic, A., Mackinder, B. A. & Herendeen, P. S. Phylogeny of the Detarioid Legume genera _Cynometra_ and _Maniltoa_ (Leguminosae). _Syst. Bot._ 42,
670–679 (2017). Article Google Scholar * Wieringa, J. J., Mackinder, B. A. & van Proosdij, A. S. J. _Gabonius_ gen. nov. (Leguminosae, Caesalpinioideae, Detarieae), a distant cousin of
_Hymenostegia_ endemic to Gabon. _Phytotaxa_ 142, 15–24 (2013). Article Google Scholar * Mackinder, B. A. & Wieringa, J. J. _Annea_ gen. nov. (Detarieae, Caesalpinioideae,
Leguminosae): a home for two species long misplaced in _Hymenostegia_ sensu lato. _Phytotaxa_ 142, 1–14 (2013). Article Google Scholar * Breteler, F. J. Novitates Gabonenses 56. Two
_Anthonotha_ species from Gabon transferred to _Englerodendron_ (Fabaceae, Caesalpinioideae). _Adansonia_ 28, 105–111 (2006). Google Scholar * Breteler, F. J. _Anthonotha_ and
_Isomacrolobium_ (Leguminosae, Caesalpinioideae): two distinct genera. _Syst. Geogr_. _Plants_ 78, 137–144 (2008). Google Scholar * Polhill, R. In _Phytochemical dictionary of the
Leguminosae Vol 1. Plants and their constituents_ Vol. 1 (eds FA Bisby, J Buckingham, & JB Harborne) Complete synopsis of legume genera. xlix-liv (Chapman & Hall, 1994). * Lewis, G.
R. & Schrire, B. D. _Micklethwaitia_, a new name for _Brenaniodendron_ J. Léonard (Leguminosae: Caesalpinioideae: Detarieae). _Kew Bull._ 59, 166–166 (2004). Article Google Scholar
Download references ACKNOWLEDGEMENTS We wish to thank the staff of the cited herbaria for their support during our visits and for the loan of material. Permission to reproduce photographs
was generously given by D. Cardoso, P. Cribb, E. Moll, C. Jongkind/Fauna & Flora International and X. van der Burgt. We thank D. Cardoso and Editor Xinwei Xu for their valuable comments
and help. Analyses were performed on the supercomputer Briarée from the Université de Montréal, managed by Calcul Québec and Compute Canada. This project was funded by a grant to A.B. from
the Natural Sciences and Engineering Research Council of Canada and M.d.l.E. was funded by the European Union’s Horizon 2020 research and innovation programme under the Marie
Sklodowska-Curie grant agreement No 659152 (GLDAFRICA). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Comparative Plant and Fungal Biology Department, Royal Botanic Gardens, Kew, Richmond,
TW9 3DS, UK Manuel de la Estrella, Félix Forest, Gwilym P. Lewis & Barbara A. Mackinder * Departamento de Botánica, Ecología y Fisiología Vegetal, Facultad de Ciencias, Campus de
Rabanales, Universidad de Córdoba, 14071, Córdoba, Spain Manuel de la Estrella * Department for Identification and Naming, Royal Botanic Gardens, Kew, Richmond, TW9 3AE, UK Bente Klitgård *
Tropical Diversity, Royal Botanic Garden Edinburgh, 20ª Inverleith Row, EH3 5LR, Edinburgh, UK Barbara A. Mackinder * Departamento de Ciências Biológicas, Universidade Estadual de Feira de
Santana, Av. Transnordestina s.n., Novo Horizonte, 44036-900, Feira de Santana, Bahia, Brazil Luciano P. de Queiroz * Naturalis Biodiversity Centre, National Herbarium of the Netherlands,
Darwinweg 2, 2333 CR, Leiden, The Netherlands Jan J. Wieringa * Institut de recherche en biologie végétale and Département de Sciences biologiques, Université de Montréal, 4101 Sherbrooke
est, Montréal, H1X 2B2, Canada Anne Bruneau Authors * Manuel de la Estrella View author publications You can also search for this author inPubMed Google Scholar * Félix Forest View author
publications You can also search for this author inPubMed Google Scholar * Bente Klitgård View author publications You can also search for this author inPubMed Google Scholar * Gwilym P.
Lewis View author publications You can also search for this author inPubMed Google Scholar * Barbara A. Mackinder View author publications You can also search for this author inPubMed Google
Scholar * Luciano P. de Queiroz View author publications You can also search for this author inPubMed Google Scholar * Jan J. Wieringa View author publications You can also search for this
author inPubMed Google Scholar * Anne Bruneau View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Designed the project: M.d.l.E. and A.B.
Compiled materials and generated data: M.d.l.E., J.J.W. and A.B. Analysed the data: M.d.l.E., A.B. and F.F. Wrote the paper: M.d.l.E., L.P.Q. and A.B. with contributions of F.F., B.K.,
G.P.L., B.A.M. and J.J.W. CORRESPONDING AUTHOR Correspondence to Manuel de la Estrella. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL
INFORMATION PUBLISHER'S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL
SUPPLEMENTARY INFORMATION RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation,
distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and
indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to
the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will
need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE de la Estrella, M., Forest, F., Klitgård, B. _et al._ A new phylogeny-based tribal classification of subfamily Detarioideae, an early branching clade of florally diverse
tropical arborescent legumes. _Sci Rep_ 8, 6884 (2018). https://doi.org/10.1038/s41598-018-24687-3 Download citation * Received: 22 December 2017 * Accepted: 28 March 2018 * Published: 02
May 2018 * DOI: https://doi.org/10.1038/s41598-018-24687-3 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative