Complete genome of the listeria monocytogenes strain auf, used as a live listeriosis veterinary vaccine
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT _Listeria monocytogenes_ (Lm) is a highly pathogenic bacterium that can cause listeriosis, a relatively rare food-borne infectious disease that affects farm, domestic, wild animals
and humans as well. The infected livestock is the frequent sources of Lm. Vaccination is one of the methods of controlling listeriosis in target farm animals to prevent Lm-associated food
contamination. Here we report the complete sequence of the Lm strain AUF attenuated from a fully-virulent Lm strain by ultraviolet irradiation, successfully used since the 1960s as a live
whole-cell veterinary vaccine. The _de novo_ assembled genome consists of a circular chromosome of 2,942,932 bp length, including more than 2,800 CDSs, 17 pseudogenes, 5 antibiotic
resistance genes, and 56/92 virulence genes. Two wild Lm strains, the EGD and the 10403S that is also used in cancer Immunotherapy, were the closest homologs for the Lm strain AUF. Although
all three strains belonged to different sequence types (ST), namely ST12, ST85, and ST1538, they were placed in the same genetic lineage II, CC7. DESIGN TYPE(S) sequence analysis objective •
sequence assembly objective• sequence annotation objective • genotyping by high throughput sequencing design MEASUREMENT TYPE(S) whole genome sequencing • genome assembly • sequence
annotation TECHNOLOGY TYPE(S) DNA sequencing • whole genome sequencing FACTOR TYPE(S) Organism Strain • live whole cell vaccine genome SAMPLE CHARACTERISTIC(S) Listeria monocytogenes SIMILAR
CONTENT BEING VIEWED BY OTHERS A EUROPEAN-WIDE DATASET TO UNCOVER ADAPTIVE TRAITS OF LISTERIA MONOCYTOGENES TO DIVERSE ECOLOGICAL NICHES Article Open access 28 April 2022 CORRELATION
ANALYSIS OF WHOLE GENOME SEQUENCING OF A PATHOGENIC _ESCHERICHIA COLI_ STRAIN OF INNER MONGOLIAN ORIGIN Article Open access 05 July 2024 FULL GENOME SEQUENCE FOR THE AFRICAN SWINE FEVER
VIRUS OUTBREAK IN THE DOMINICAN REPUBLIC IN 1980 Article Open access 19 January 2023 BACKGROUND & SUMMARY _Listeria monocytogenes_ (Lm), a highly pathogenic bacterium, is a well-known
causative agent of listeriosis. This infection has been proven to be one of the important life-threatening food-borne infections for many species, such as humans, wildlife and farm animals,
wild birds, and poultry, being a major concern for both veterinary and public health worldwide1,2,3,4,5,6,7. Lm pathogen was first recognized as a pathogen in the 1920s during an outbreak in
laboratory animals, such as rabbits1,2,8. However, research interest in _Listeria_ has risen significantly since the 1980s, when the critical role of Lm as the etiologic agent responsible
for sporadic cases and numerous outbreaks of human listeriosis was established. These episodes were strictly associated with the consumption of contaminated foods, mainly in Western Europe,
Canada, the USA, and Africa2,3,6,9,10,11,12,13,14,15. Listeriosis is a typical primary zoonotic infection that can be also transmitted vertically in infected pregnant animals and humans,
crossing the blood-brain and placental barriers and leading to an invasion into the central nervous system and fetus infection2,3,5,8,9,14,16,17,18,19,20,21,22. This ubiquitous Gram-positive
bacterium can produce in infected mammals a systemic infection leading to bacteremia with serious complications, such as septicemia, meningitis and other central nervous systems (CNS)
pathology. Moreover, the pathogen can cause severe focal infections and adverse maternal–neonatal outcomes, including abortion, stillbirth, and preterm birth. Overall, neonatal listeriosis
accounts for more than 20% of fatal cases2,3,5,8,19,20,21,22. Pregnant women, the elderly, and immunocompromised individuals have the highest risk of developing complications and mortality
from listeriosis2,3,5,8,13,19,20. Infected farm animals are one of the sources of Lm5,6,7,10. Fecal shedding of Lm was found in dairy cattle (46.3%), beef cattle (30.6%) and sheep herds
(14.2%)10. Vaccination is considered one of the methods of choice to control listeriosis in target farm animals to prevent the spread of this infection and Lm-associated food contamination,
especially for meat and dairy products4,6,10,17,18,23,24. Nevertheless, inactivated, live avirulent, or fully virulent Lm strains have been reported to fail in inducing the protective immune
response against animal listeriosis6. The attenuated Lm strain AUF has been successfully used for almost 50 years in some regions of the former USSR as a live whole-cell vaccine (LWCV)
against listeriosis in farm animals. The Lm strain AUF derives from the fully virulent Lm strain ‘A’ isolated in 1965 from the pathological material of the brain of an ewe with
neurolisteriosis23,24. The attenuation was done by a combination of 17 repeated exposures of the original strain ‘A’ to Ultraviolet radiation (UVR)23,24. The selected Lm strain AUF
demonstrated low virulence in animal models (outbred white mice and rabbits) and pronounced immunogenicity in target farm animals (sheep, goats, cows, and pigs)23,24. Since the late 1960s,
the commercial LWCV based on the Lm strain AUF has been manufactured for the prevention of listeriosis in animal husbandry. However, the genome of the Lm strain AUF has not been sequenced.
It is critical to identify the possible genetic markers contributing to the attenuation and residual virulence of this particular Lm strain, which has been used as a commercial LWCV for a
long time. In order to elucidate the basic genomic features of this unique strain, two platforms - Illumina HiSeq 2500 (Illumina Inc., USA) and Nanopore MinION (Oxford Nanopore, UK),
intended for short-read and long-read sequencing strategies, respectively, were used in this study in parallel. After quality filtering following the trimming of ‘raw’ massive data, the _de
novo_ hybrid assembly of the complete genome sequence of the Lm strain AUF represented by a single chromosome 2.94 Gb in size, was successfully obtained. Structural and functional analysis
of the Lm strain AUF genome25 showed that the assembled complete Lm strain AUF chromosome contained 2,963 genes, including 2,874 CDSs, 17 pseudogenes, 89 RNA genes, 6 rRNA (5 S, 16 S, 23 S),
67 tRNA and 4 non-coding RNA, 5 antibiotic resistance genes (_fosX_, _mprF_, _lin_, _norB_ and _sul_), 56 of 92 genes associated with Lm virulence26,27 and immunogenicity factors26,27,
including such key virulence factors as _inlA_ and _inlB_ which encoded internalin A (InlA) and internalin B (InlB) responsible for binding the host cell receptors E-cadherin and Met,
respectively; six genes of the pathogenicity island LIPI-1, _plcA_, _hly_, _mpl_, _actA_, _plcB_ and _prfA_, which encoded the transcriptional regulator positive regulator factor A (PrfA)
controlling the expression of both inlAB locus and LIPI-128,29,30,31,32 etc.). Additionally, the five-gene stress survival islet (SSI-1, _lmo0444_, _lmo0445_, _lmo0446_, _lmo0447_ and
_lmo0448_), which is known to confer resistance to environmental stress, such as low pH, high osmolarity, bile and nisin33, was annotated in the Lm strain AUF suggesting the possible
contribution of these genes to the adaptation of the Lm strain AUF to the relevant conditions. Unfortunately, the culture and sequence of the parental Lm strain ‘A’ isolated more than 50
years ago, is not currently available. In fact, analyses for mutations that might have been induced by the UVR exposure can therefore not be accurately confirmed but rather inferred from
closely related fully virulent Lm strains. For this reason, the whole-genome sequence of the Lm strain AUF was compared with nine assembled Lm strains available in GenBank, which had been
earlier isolated from different animals with listeriosis, including the referent fully virulent Lm standard strains, the Lm strain FDAARGOS_607, and the Lm strain EGD-e strains approved by
the US Food and Drug Administration and the European Consortium, respectively34,35,36,37. Additionally, the genomes of the Lm strain 10403S and the Lm strain EGD, which have utilized
together with the Lm strain EGD-e to analyse the Lm virulence38, were selected. We found two groups of loci, which were identified in: (i) both the Lm strain AUF and fully virulent reference
strains (virulence (56/73, 76.7%), antibiotic resistance (5/5, 100%), SSI-1 (5/5, 100%), Lm Genomic Islands (2/34, 5.9%), motility (n = 29/31, 93.5%); (ii) only in the fully virulent Lm
strains, but not in the Lm strain AUF (virulence (17/73, 23.3%), cadmium resistance (n = 2/2, 100%), SSI-2 (n = 2/2, 100%), Lm Genomic islands (n = 32/34, 94,1%), motility (n = 2/31, 6,5%).
Moreover, marked polymorphisms were instantly detected in both nucleotide sequences and allelic profiles of the Lm strain AUF major virulence genes, such as _pfrA_, _hly_, _inlA_ and _inlB_
which were present in other fully and weakly virulent Lm strains. Importantly, the Lm strain AUF was not absolutely genetically close (3,690 SNPs) to the Lm strain EGD; the Lm strain AUF
demonstrated no point mutation in the transcriptional regulator _prfA_ typical for the Lm strain EGD compared with the Lm strain EGD-e as G145S38. Surprisingly, the Lm strain AUF was
genetically close (148 SNPs) to the Lm strain 10403S, which was successfully utilized for the development of attenuated Lm-based cancer vaccine vectors, exploring the Lm unique life cycle
and ability to induce robust Cytotoxic T lymphocytes (CTLs) immune responses39,40,41,42,43,44. Nevertheless, contrary to the Lm vectors used in the Lm cancer vaccination platforms which
contained truncated virulence genes _prfA_, _actA_ and _plcB_39,40,41,42,43,44, both Lm strains, the Lm strain AUF and the Lm strain 10403S, demonstrated the presence of the intact variants
of the relevant genes. The Lm strain AUF was markedly distinct from other Lm reference strains, such as the Lm strain EGD-e (31,385 SNPs), the Lm strain NTSN (141,219 SNPs), the Lm strain
FDAARGOS_607 (144,871 SNPs), the Lm strain UKVDL9 (178,505 SNPs), the Lm strain UKVDL4 (178,552 SNPs), the Lm strain UKVDL7 (180,314 SNPs), the Lm strain 4/52-1953 (182,819 SNPs) and the Lm
strain FSL-J1-158 (196,422 SNPs). The data presented here is the first report highlighting the genome loci which could be either directly or indirectly involved in the attenuation (LIPI-3,
_srtB_, _agrC_, _vip_, _gltB_, _gltA_, _aut_IVb_, metal resistance genes and Lm Genomic Islands-associated genes) and residual virulence (LIPI-1, _plcA_, _hly_, _mpl_, _inlA_, _inlB_,
_prfA_, _hly_, _actA_ and _plcB_) of the single Lm strain AUF with a long history of application as an effective veterinary LWCV against listeriosis. Our data could also serve as the basis
for unravelling the mechanisms of virulence and pathogenicity in Lm. The results of this study will be useful for improving our knowledge of bacterial vaccinology overall and developing of a
new generation of vaccines against listeriosis in farm animals. This is critical for animal health and welfare, food safety, and human public health worldwide. METHODS BACTERIAL STRAIN The
Lm strain AUF was obtained from the Collection of Microorganisms of the Department for Microbiology and Biotechnology, Saratov State University of Genetics, Biotechnology and Engineering
named after N.I. Vavilov, Saratov, Russia. The Lm strain AUF was stored in lyophilized form. The Lm strain AUF was routinely cultivated on Tryptone Soy Yeast Extract Agar (TSYE Agar) (Merck,
EU) overnight prior to the experiments as described45,46. DNA EXTRACTION FOR SHORT & LONG READS SEQUENCING Genomic DNA was extracted from the lysate of the Lm strain AUF culture, using
a commercial DNA DNeasy Blood & Tissue Kit (Qiagen, Germany) according to the manufacturer’s instructions as described45,46. The final DNA concentration was measured using a
spectrophotometer from BioRad (Bio-Rad, USA). WHOLE GENOME SEQUENCING The isolated DNA from the Lm strain AUF strain was used in two parallel sequencing platforms, Illumina HiSeq 2500
(Illumina Inc., USA) and Nanopore MinION (Oxford Nanopore, UK). For this purpose, the preparation of DNA libraries was conducted using either Nextera XT DNA Library Preparation Kit (Illumina
Inc., USA) or Nanopore Kit SQK-LSK109 (Oxford Nanopore, UK), respectively, as we described recently45,46. In the first case, the DNA was sequenced commercially as 250-bp single-Read
(Illumina Inc., USA) at Genoanalytica LLC (https://www.genoanalytica.ru/, Moscow, Russia). A FLO-MIN-106 R9.4 Flow cell (Oxford Nanopore Technologies, Oxford, UK) was routinely used to
perform sequencing with the MinION and the MinKNOW software as recommended (https://nanoporetech.com/). GENOME ASSEMBLING AND ANNOTATION Unicycler v0.4.9 with default parameters
(https://github.com/rrwick/Unicycler) was used to generate a hybrid high-quality _de novo_ assembly of the Lm strain AUF strain genome47. The annotation of chromosome was performed by the
NCBI Prokaryotic Genome Annotation Pipeline (PGAP) with default parameters on the submission portal of NCBI GenBank (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/)48. Construction of
the phylogenetic tree based on the Lm strain AUF and other _Listeria spp_. strain whole genomes was conducted with the online tool REALPHY 1.13 using default parameters
(https://realphy.unibas.ch/realphy/)49 and MEGA-7 software50 by the maximum likelihood method for the generation of the phylogenetic tree based on the internalin genes. Antibiotic resistance
genes were identified using CARD (https://card.mcmaster.ca/analyze/rgi) and the Antibiotic Resistance scheme of the Institute Pasteur Bacterial Isolate Genome Sequence Database (BIGSdb
Version 1.42.0) (https://bigsdb.pasteur.fr/listeria/). Comparative analysis of the CDSs in the Lm strain AUF strain relative to the Lm reference strains was performed using «Proteome
Comparison Service» tool on The Bacterial and Viral Bioinformatics Resource Center (BV-BRC) platform51. Visualization of the linear map of the Lm strain AUF genome was generated using the
online tool Proksee (https://proksee.ca/)52. Genes encoding virulence factors in the Lm strain AUF and other Lm strains were found using the BIGSdb-Lm database
(https://bigsdb.pasteur.fr/listeria/). Identification of allele profiles of the Lm strain AUF and reference Lm strains whole genome and Multilocus sequence typing (MLST) was also performed
using the BIGSdb-Lm database (https://bigsdb.pasteur.fr/listeria/). Comparative analysis of pseudogenes in the Lm strain AUF and other Lm strains was performed using BLAST
(https://blast.ncbi.nlm.nih.gov/Blast.cgi). A PANEL OF LM REFERENCE STRAINS We compared the annotated completed genomes of ten Lm strains available in the NCBI GenBank
(https://www.ncbi.nlm.nih.gov/). The relevant whole genome sequences were downloaded: the Lm strain FDAARGOS_60753, the Lm strain EGD-e54, the Lm strain EGD55, the Lm strain FSL-J1-15856,
the Lm strain NTSN57, the Lm strain UKVDL958, the Lm strain UKVDL459, the Lm strain 4/52-195360, the Lm strain UKVDL761 and the Lm strain 10403S62. Additionally, the genomes of 12 strains of
other _Listeria spp_. were used for the construction of a phylogenetic tree in order to demonstrate the phylogenetic relationships between the Lm strain AUF and other _Listeria_, such as:
the _Listeria grayi_ strain NCTC1081263, the _Listeria valentina_ strain CLIP 2019/0064264, the _Listeria kieliensis_ strain Kiel-L165, the _Listeria floridensis_ strain FSL S10-118766, the
_Listeria fleischmannii_ strain 199167, the _Listeria rustica_ strain FSL W9-058568, the _Listeria newyorkensis_ strain CMB19106369, the _Listeria cornellensis_ strain FSL F6-096970, the
_Listeria rocourtiae_ strain CECT 7972 Ga0244616_10171, the _Listeria booriae_ strain FSL A5-028172, the _Listeria riparia_ strain FSL S10-120470, the _Listeria phage_ strain A11873. DATA
RECORDS The completed whole genome sequence of the Lm strain AUF has been deposited in GenBank with the accession number CP048400.147. All of the reads for the Lm strain AUF genome have been
deposited in the NCBI Sequence Read Archive under the accession numbers SRR25210281 for Oxford Nanopore data74 and SRR25180708 for Illumina HySeq 2500 data)75. TECHNICAL VALIDATION Before
starting the genome assembly, raw reads were prepared using an automated AfterQC script to remove low-quality reads and Illumina adapters in default settings for short reads, as well as to
remove adapters using Filtlong v0.2.1 software. To filter long reads by quality obtained on the Oxford Nanopore platform, we used the Filtlong script (https://github.com/rrwick/Filtlong)
with the filter parameters --min_length 1000 --keep_percent 90. Trimming and searching for Nanopore adapter sequences in long reads was performed using the Porechop script according to the
recommended parameters (https://github.com/rrwick/Porechop). The _de novo_ assembly using both short and long reads, obtained from both platforms, Illumina and Nanopore MinION, generated the
Lm strain AUF circular chromosome of 2,942,932 bp in size with a GC content of 37.98%. To clarify the taxonomic affiliation of the assembled Lm strain AUF genome to other _Listeria spp_.
representatives, we constructed a phylogenetic tree based on the whole genomes available in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/). For this purpose, we selected the reference
genomes of 15 different _Listeria spp_. and 9 Lm strains associated with listeriosis in animals. The Lm strain AUF formed a separate branch with representatives of the Lm only, but not with
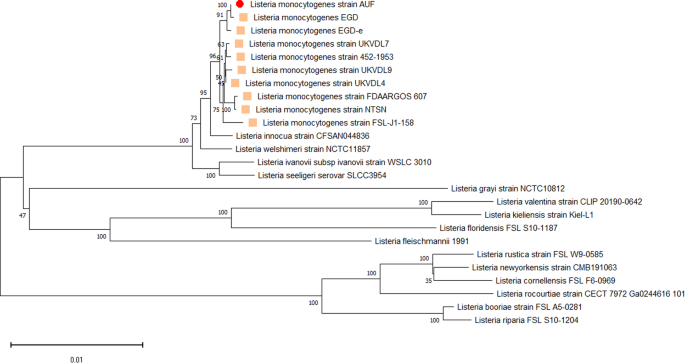
other _Listeria spp_. reference bacteria strains, which proved its genetic affiliation to the indicated Lm species (Fig. 1). Importantly, the Lm strain AUF visualized in a single clade with
the fully virulent Lm strain EGD, which was isolated in 1924 in the United Kingdom from a guinea pig infected with biomaterial derived from a rabbit with listeriosis during the disease
outbreak in laboratory animals1,2,8. The data obtained recognized the EGD as the closest homolog to the Lm strain AUF, in contrast to other Lm representatives, including both the Lm strain
EGD-e and the Lm strain FDAARGOS_607 (Fig. 1). The Lm strain EGD-e was assigned as the second homolog for the Lm strain AUF located in the same cluster with the Lm strain AUF and the Lm
strain EGD strains (Fig. 1). The Lm strain AUF was also distinguished from the Lm 4/52-1953, had also been isolated on the territory of the former USSR, although a decade earlier46. In fact,
recently phylogenetic analysis based on whole genomes of Lm strains (n = 257) available in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/) showed that these two Lm strains belonged to
different Clusters, the Lm strain AUF to the Cluster II, represented by genetic lineages I and II, whereas the Lm strain 4/52-1953 - to the Cluster I, formed by genetic lineage III only46.
Interestingly, the Lm strain AUF was phylogenetically closer to the Lm strain 10403S derived from the clinical sample of a human with listeriosis46. Both strains, the Lm strain AUF and the
Lm strain 10403S, formed a single Cluster II46. The _prfA_-, or _act_A- or _plcB_- defective derivatives of the Lm strain 10403S were reported to be safe live attenuated Lm vectors for the
development of cancer immunotherapy vaccines39,40,41,42,43,44. Through MLST, based on the sequences of seven housekeeping genes (_abcZ_, _bglA_, _cat_, _dap_, _dat_, _ldh_ and _lhkA_), we
found that the Lm strain AUF together with the closest homologs, the fully virulent reference strains, the Lm strain EGD, the Lm strain EGD-e and the Lm strain 10403S, belonged to the same
genetic lineage II, although these strains sourced from different hosts, such as sheep, rabbit and human, respectively (Table 1). Moreover, the Lm strain AUF, the Lm strain EGD and the Lm
strain 10403S corresponded to the identical clonal complex (CC) CC7, contrary to the Lm strain EGD-e which was assigned to CC9. Nevertheless, the Lm strain AUF, the Lm strain EGD and the Lm
strain 10403S related to the different sequence types (ST), ST1538, ST12 and ST85, respectively, although all three strains demonstrated six of seven identical alleles (_abcZ_, _bglA_,
_cat_, _dap_, _dat_, and _lhkA_) with polymorphism in only a single allele, _ldh_ (Table 1). We found only one individual allele, _lhkA_, which was identical for the Lm strain AUF, the Lm
strain EGD, the Lm strain 10403S and the Lm strain EGD-e. No identical alleles were revealed between the Lm strain AUF and two ovine fully virulent Lm strains belonging to the genetic
lineage I, the Lm FDAARGOS_607 and the Lm strain NTSN, and the ovine, bovine, porcine, equine and caprine Lm strains, the Lm strain UKVDL9, the Lm strain UKVDL4, the Lm strain 4/52-1953, the
Lm strain UKVDL7, the Lm strain FSL-J1-158, the Lm strain the Lm strain UKVDL7 and the Lm strain of the genetic lineages III-IV belonging to different STs, ST1194, ST1069, ST201, ST1140 and
ST563 of clonal complexes ST1194, ST1069, CC69, CC1070 and ST563 (Table 1). Comparative analysis of the coding regions using BLASTP51 proved the marked discrimination of the Lm strain AUF
genome from the other ten representative Lm strains (Fig. 2). As expected, the Lm strain AUF showed the highest homology of coding regions with the Lm strain 10403S, the Lm strain EGD, a few
somewhat lower one with the Lm strain EGD-e one, and much lower one with other Lm reference strains compared. Comparative analysis of the whole genomes of the Lm strain AUF versus nine Lm
reference strains of zoonotic source on the BV-BRC platform (https://www.bv–brc.org/) showed 100% homology in more than 90% CDSs in the Lm strain EGD strain, approximately 50% in the Lm
strain EGD-e and only 12–14% in other Lm genomes (Table 2). Similarly, the highest homology in CDSs ranging from 20–79% demonstrated in the majority of Lm strains except both closest
homologs. Also, approximately 200 CDSs were present in only the Lm strain AUF and absent in all other strains except the Lm strain EGD. About 100 CDSs were annotated in the Lm strain AUF but
not in the Lm strain EGD76. These Lm strain AUF-specific genes were located on the Lm strain AUF chromosome as several compact regions, which were visualized as extended insertions in the
Lm strain AUF compared with other reference Lm strains used, including both closest homologs, the Lm strain EGD and the Lm strain EGD-e, and the Lm strain 10403S as well (Fig. 2). The
majority of these Lm strain AUF-specific genes encoded uncharacterized hypothetical proteins, _Listeria_ CRISPR-associated proteins (Cas1/Cas2/Cas9/Csn2)77, phage proteins, Type I
restriction-modification system components, and in contrast to some Lm strains, GNAT family Acetyltransferase, Gp45 protein, 4-hydroxy-2-oxoglutarate aldolase (EC 4.1.3.16),
2-dehydro-3-deoxyphosphogluconate aldolase (EC 4.1.2.14), transcriptional regulator, ADP-ribosyl glycohydrolase, putative EsaC protein analog (_Listeria_ type 3), cassette chromosome
recombinase B, and mobile element proteins. Bacteriophage A118 genes which have been known as _Listeria_ phage A118 (NCBI Acc. number: NC_003216.1), and initially hosted in the Lm strain
EGD-e78,79,80,81, were successfully detected in the Lm strain AUF and all other Lm strains used in the current study except the Lm strain EGD76. Comparison of the whole genomes of the Lm
strain AUF with the Lm strain 10403S sourced from human sample using the similar BV-BRC platform (https://www.bv–brc.org/) demonstrated 100% homology for about 93% CDSs, 80 – 99% identity
for slightly more than 3% CDSs and less than 1% in CDSs ranging from 20-79% (Table 2). Only 93 CDSs were annotated in the Lm strain AUF. These were absent in the Lm strain 10403S76.
Basically, there were the same Lm strain AUF-specific genes which encoded uncharacterized hypothetical proteins and phage proteins76. Additionally, we found (Table 3) in the Lm strain AUF
approximately 60% (56/92) of genes that have been recently recognized as those involved in Lm virulence26. This amount was 7-10% less than that revealed in both fully virulent Lm strains of
the genetic lineage I, the Lm strain NTSN (62/92, 67.4%) and the Lm strain FDAARGOS_607 (64/92, 69.6%). However, the number of virulence genes in the Lm strain AUF was certainly higher than
that in the Lm strains of the genetic lineages III-IV, up to 1.3 - 3.9 times, for instance, in the Lm strain UKVDL7 (14/92, 15.2%) and the Lm strain 4/52-1953 (41/92, 44.6%), respectively.
In fact, the Lm strain AUF demonstrated almost similar characteristics in this regard compared with the other two Lm strains of zoonotic origin, the Lm strain EGD and the Lm strain EGD-e
(57/92, 61.9%), and with the Lm human strain 10403S (57/92, 61.9%), also belonging to the genetic lineage II. Importantly, 14 specific loci were identified in both Lm strains of the lineage
I, the Lm strain FDAARGOS_607 and the Lm strain NTSN (Table 1), but not in the Lm strain AUF25. There were the following genes: of LIPI-3, the additional sub-lineage pathogenicity island
encoding listeriolysin S (LLS), a hemolytic toxin, a bacteriocin31,32 (LIPI3_llsB (LMOf2365_1116), LIPI3_llsD (LMOf2365_1118), LIPI3_llsY (LMOf2365_1117), LIPI3_llsG (LMOf2365_1113),
LIPI3_llsH (LMOf2365_1114), LIPI3_llsP (LMOf2365_1119), LIPI3_llsX (LMOf2365_1115), LIPI3_llsA (LMOf2365_1112a); _srtB_ (lmo2181), encoded the sortase B peptidoglycan-anchored protein
induced in low iron conditions and involved with other sortases in many aspects of pathogenesis, from biofilm formation, adhesion, and immune suppression to bacterial loads and
lethality82,83; _agrC_ (lmo0050), responsible for the expression of AgrC, an integral membrane protein, a member of the class 10 receptor histidine protein kinases which are identified in Lm
and involved in a quorum sensing system for the regulation of virulence (internalization, toxins) and biofilm84,85,86,87; _vip_ (lmo0320), encoded vegetative insecticidal proteins (VIPs),
toxins, during the vegetative growth phase by different bacterial species88; _gltB_ (LMOf2365_2741), which encodes the major subunit of the glutamate synthase; _gltA_ (LMOf2365_2740),
citrate synthase gene, a critical mediator of site-specific fitness of pathogens during infection due to its influence on metabolic flexibility89; aut_IVb (LMOF2365_RS00075), which encodes
autolysin, a member of a group of bacterial hydrolases involved in Lm invasion90,91,92. Less difference was noted between the Lm strain AUF and the Lm strain EGD. The latter strain contained
only two additional loci, _srtB_ (lmo2181) and _agrC_ (lmo0050) which were absent in the Lm strain AUF, but were found in the fully virulent strains, such as the Lm strain EGD-e, the Lm
strain NTSN, and the Lm strain FDAARGOS_607. We identified three loci, _srtB_ (lmo2181), _agrC_ (lmo0050), and _vip_ (lmo0320) in the Lm strain EGD-e and in the Lm strain 10403S but not in
the Lm strain AUF. The Lm strain NTSN had 13 additional loci which were not found in the Lm strain AUF. The same loci were present in the Lm strain FDAARGOS_607, except for aut_IVb
(LMOF2365_RS00075), and absent in the Lm strain AUF. Only two specific loci (_agrA_ (lmo0051) and _comK_ (LMOf2365_2303)) were revealed in the Lm strain AUF, being absent in the Lm strain
10403S25. The locus _prfA_ (lmo0200), which encodes the transcriptional factor PfrA, the master regulator of Lm major virulence genes39,93,94,95, was identified in the Lm strain AUF
similarly with other Lm reference strains25. Remarkably, the Lm strain AUF PrfA demonstrated no 2 amino acid changes typical for the Lm strain EGD (Serine → Glycine, S145G; Tyrosine →
Cysteine, Y229C), resulting in the constitutive overexpression in the Lm strain EGD of several major virulence genes39 although the marked _prfA_ nucleotide sequence polymorphism96. Multiple
sequence alignment of the Lm strains AUF PrfA protein versus other Lm reference strains showed its identity with those in the Lm strains, the Lm strain EGD-e, the Lm strain FDAAGROS_607,
the Lm strain NTSN, the Lm strain UKVDL4 and the Lm strain 10403S97, thus supposing the possible expression in the Lm strain AUF of the PrfA-regulated virulence genes comparable to the Lm
strain EGD-e and other virulent Lm strains as well. The locus _hly_ (lmo0202) encoded the pore-forming toxin listeriolysin-O (LLO), allowing Lm to escapes the phagosome to avoid lysosomal
killing43,98 was also identified in the Lm strain AUF and other Lm reference strains96,99. The alignment of the Lm strain AUF LLO protein was almost identical to those in the Lm strain EGD,
the Lm strain EGD-e and the Lm strain 10403S, although differed from all the other Lm strains by a single amino acid substitution of one Serine to Lysine, S523K. Also, there were 3 other
amino acid substitutions, namely: (i) Threonine to Asparagine, T309N in the Lm strain AUF and other strains except the Lm strain FSL-J1-158; (ii) Asparagine, N in position 31 in the Lm
strain AUF, the Lm strain EGD, the Lm strain EGD-e, the Lm strain 10403S, the Lm strain 4/52-1953, the Lm strain FDAAGROS_607 and the Lm strain NTSN instead of either Histidine, H in the Lm
strain UKVDL4, the Lm strain UKVDL7 and the Lm strain UKVDL9 or Glutamine, Q in the Lm strain FSL-J1-158; (iii) Serine, S in the position 35 in the Lm strain AUF, the Lm strain EGD, the Lm
strain EGD-e, the Lm strain 10403S, the Lm strain 4/52-1953 and the Lm strain FSL-J1-158 instead of Leucine, L in the Lm strain FDAAGROS_607, the Lm strain NTSN, the Lm strain UKVDL4, the Lm
strain UKVDL7 and the Lm strain UKVDL999. Also, 6 specific loci (_agrA_ (lmo0051), _ami_ (lmo2558), _aut_ (lmo1076), _inlG_ (lmo0262), _tagB_ (lmo1088), and _inlL_ (LMON_RS10535), were
found only in the Lm strain AUF and were not identified in the Lm strain FDAARGOS_607, while only a single locus, _inlC_ (lmo1786) was absent in the Lm strain EGD (locus identified in the Lm
strain FDAARGOS_607). We found in the Lm strain AUF two loci, _inlD_ (LMON_RS01345) and _comK_ (LMOf2365_2303) that were not recognized in the Lm strain EGD-e (both of them were also
identified in the Lm strain FDAARGOS_607). At least seven loci, _prsA2_ (lmo2219), _fbpA_ (lmo1829), _ami_ (lmo2558), _aut_ (lmo1076), _inlG_ (lmo0262), _inlL_ (LMON_RS10535), and _tagB_
(lmo1088) were undetectable in the Lm strain NTSN strain while present in the Lm strain AUF25. There were 12 out of 56 (21.5%) identical loci present in the Lm strain AUF and the Lm strain
FSL-J1-158 of the genetic lineage IV, and from 13 out of 56 (23.2%) to 39 out of 56 (69.6%) in the Lm strains of the genetic lineage III. Apparently, either all or some of these loci could
be potentially either directly or indirectly involved in the mechanisms of residual virulence of the Lm strain AUF. Furthermore, 14 genes encoding biosynthesis of members of the internalin
multigene family100, internalins (_inlA_ (lmo0433), _inlB_ (lmo0434), _inlC_ (lmo1786), _inlC2_ (LMON_RS01340), _inlD_ (LMON_RS01345), _inlE_ (lmo0264), _inlF_ (lmo0409), _inlG_, _inlH_
(lmo0263), _inlI_ (lmo0333), _inlJ_ (lmo2821), _inlK_ (lmo1290), _inll_ and _inlP_ (lmo2470) were found in the Lm strain AUF and the majority of the Lm strains belonging to the genetic
lineages I-II, while rarely detected in the representatives of the genetic lineages III-IV (Table 4). Comparative analysis of the main internalin genetic characteristics showed that the
majority of genes and the relevant allelic profiles of the Lm strain AUF were almost identical to those found in the Lm strain EGD and the Lm strain 10403S strains. The _inlC_ was present in
the Lm strain AUF and absent in the Lm strain EGD. The difference between the Lm strain AUF and the Lm strain 10403S was found only in the alleles for the _inlB_. Only a single identical
allele (_inlG_) was present in both the Lm strain AUF and the Lm reference strain EGD-e. No identical allele profiles for these genes were found between the Lm strain AUF and both Lm strains
of the genetic lineage I, the Lm strain FDAARGOS_607, and the Lm strain NTSN. The Lm strain AUF, likewise other Lm strains of the genetic lineage II, the Lm strain 10403S, the Lm strain
EGD, and the Lm reference strain EGD-e, showed a certain diversity in internalin gene compositions versus the Lm strains of the genetic lineages III-IV, similarly to other
reports100,101,102. No internalin genes, including _inlA_ and _inlB_, in the form of either pseudogenes or truncated variants were present in the Lm strain AUF and the Lm strain 10403S, the
Lm strain EGD, and the Lm strain EGD-e unlike the Lm reference strains of both these genetic lineages (Table 4). However, there was a marked polymorphism in the multiple sequence alignment
of _inlA_96,103 and _inlB_96,104. The Lm strain AUF together with the Lm strain EGD and the Lm strain 10403S differed from other Lm reference strains by the presence of 3 major amino acid
substitutions in InlA protein as T51A (Alanine instead of Threonine), S187N (Asparagine instead of Serine), and A594P (Proline instead of Alanine). Further, we found in the same protein of
the Lm strain AUF and 3 other phylogenetically close Lm strains, the Lm strain EGD, the Lm strain EGD-e and the Lm strain 10403S at least 10 amino acid substitutions, such as R3K (Lysine
instead of Arginine), S142T (Threonine instead of Serine), N474S (Serine instead of Asparagine), S476P (Proline instead of Serine), Y530H (Histidine instead of Tyrosine), T648S (Serine
instead of Threonine), T664A (Alanine instead of Threonine), N738D (Aspartate instead of Asparagine), I781L (Leucine instead of Isoleucine) and V790M (Methionine instead of Valine)97,103.
More differences were found for _inlB_ alignments. The InlB amino acid sequences of the Lm strain AUF and both the Lm strain EGD and the Lm strain 10403S had a single amino acid substitution
in position 396 resulting in a change of Alanine to Threonine (A396T). The Lm strain AUF demonstrated 23 either single or double substitutions as amino acid changes in the InlB identical to
those found in three genetically related Lm strains, the Lm strain EGD, the Lm strain EGD-e and the Lm strain 10403S, which were not identified among other Lm reference strains96,104. There
were such changes as: L176I (Isoleucine instead of Leucine), S205A (Alanine instead of Serine), S246P (Proline instead of Serine), T/M251S (Serine instead of Threonine/Methionine), I262T
(Threonine instead of Isoleucine), I291T (Threonine instead of Isoleucine), S373N (Asparagine instead of Serine), M387V (Valine instead of Methionine), E446K (Lysine instead of Glutamate),
I479M (Methionine instead Isoleucine), I483R (Arginine instead Isoleucine), P486S (Serine instead Proline), A489S (Serine instead of Alanine), TL501-502KH (Lysine-Histidine instead of
Threonine-Leucine), D533G (Glycine instead of Aspartate), I555K (Lysine instead of Isoleucine), IQ558-559TR (Threonine-Arginine instead of Isoleucine-Glutamine), GN568-569AG (Alanine-Glycine
instead of Glycine-Asparagine), V578A (Alanine instead of Valine), S580N (Asparagine instead of Serine), W584R (Arginine instead of Tryptophan), T594K (Lysine instead of Threonine),
RT599-600CQ (Cysteine-Glutamine instead of Arginine-Threonine). As expected, phylogenetically, all the internalins of the Lm strain AUF formed a single branch with only the Lm strain EGD and
the Lm strain 10403S (Fig. 3a,b,e,f,h,j,k,l), being additionally clustered with the Lm strain EGD-e (Fig. 3c,d) but not with other Lm strains (Fig. 3a-l). Moreover, in the Lm strain AUF
some of the internalins, _inlA_, _inlB_, _inlC2_, _inlD_, _inlE_ and _inlG_ were found as internalin gene clusters105, inlAB and inlC2DEG, while others, _inlC_, _inlF_, _inlH_, _inlI_,
_inlJ_, _inlK_, _inlL_ and _inlP_ were located on the chromosome outside any cluster (Fig. 4.1) similarly with the majority of the Lm reference strains (Fig. 4.2, 4.3, 4.4, 4.5, 4.6, 4.7,
4.8, 4.9, 4.10, 4.11). Two identical internalin gene clusters were identified in the Lm strain EGD (Fig. 4.3) and in the Lm strain 10403S (Fig. 4.11). The internalin gene cluster inlAB
relevant to the Lm strain AUF was also revealed in the Lm strain EGD-e and almost all the Lm reference strains (Fig. 4.4, 4.5, 4.7, 4.9, 4.10) except the Lm strain UKVDL7 (Fig. 4.8). The
second internalin cluster inlC2DEG identified in the Lm strain AUF, the Lm strain EGD and in the Lm strain 10403S was only partially presented in other Lm reference strains as either inlC2DE
in the Lm strain NTSN (Fig. 4.5) or inlDH in the Lm strain FDAARGOS_607 (Fig. 4.4), or inlEH in the Lm strain UKVDL4 (Fig. 4.7), or the Lm strain UKVDL7 (Fig. 4.8), or inlDEH in the Lm
strain UKVDL9 (Fig. 4.9). The position of both internalin gene clusters was on the relevant Lm chromosome region on about 2.0 Mbp (Fig. 4.1), while for the Lm strain EGD-e, the Lm strain
EGD, the Lm strain NTSN, the Lm strain UKVDL4, the Lm strain UKVDL9 and the Lm strain 10403S these loci were found within 0.3 – 0.5 Mbp (Fig. 4.2, 4.3, 4.5, 4.7, 4.9, 4.11) or about 0.9 –
1.1. Mbp for the Lm strain FDAARGOS_607 (Fig. 4.4), or 0.5 Mbp for the Lm strain FSL-J1-158 and the Lm strain 4/52-1953 (Fig. 4.6, 4.10). Probably, some of these genes can encode for the
products potentially involved in the “residual virulence” of the Lm strain AUF, resulting in occasional vaccine-related adverse effects in vaccinated animals. Five antibiotic resistance
genes were annotated in the Lm strain AUF related to _Listeria_ antibiotic resistance (_fosX_ (lmo1702), fosfomycins106; _mprF_ (lmo1695), cationic antimicrobial peptides107; _lin_
(lmo0919), lincosamides108; _norB_ (lmo2818), quinolones109; and _sul_ (lmo0224), sulfonamides109), which were similar to those existing in two fully virulent Lm strains of the lineage I,
the Lm strain FDAARGOS_607 and the Lm strain NTSN, and both homologs, the Lm strain EGD, the Lm strain EGD-e and the Lm strain 10403S (the lineage II), as well as in the Lm strain 4/52-1953
isolate of the genetic lineage III (Table 3). Four of the same five genes, _fosX_, _mprF_, _lin_ and _sul_, were found in the Lm strain UKVDL4. Only some of these genes were annotated in
other Lm strains of the lineages III-IV, the Lm strain UKVDL7 (n = 1), the Lm strain UKVDL9 (n = 2), and the Lm strain FSL-J1-158 (n = 1)25. No metal resistance genes were found in the Lm
strain AUF using BIGSdb-Lm database (https://bigsdb.pasteur.fr/listeria/)25. Similarly, no relevant genes were revealed in the majority of other Lm reference strains while cadmium resistance
genes, _cadA_ and _cadC_ 110,111, were identified in the Lm strain EGD-e and the Lm strain FDAARGOS_607 (Table 3)25. We found no significant difference in the number of genes involved in
motility in the Lm strain AUF compared with other Lm strains. However, the number of Lm Genomic Islands-associated genes found in the Lm strain AUF (n = 2) was markedly (18 times) lesser
those present exclusively in the Lm strain FDAAGROS_607 (n = 36). Quantitatively, the number of genes of SSI-1 (n = 5) in the Lm strain AUF strain was the same only with the closest
homologs, the Lm strain EGD, the Lm strain EGD-e and the Lm strain 10403S, and exceeded those which were annotated for other reference strains independently from their genetic lineage,
including both fully virulent strains, the Lm strain FDAARGOS_607, and the Lm strain NTSN, by 2.5 - 5 times. In contrast to the Lm strain AUF, only a single gene, SSI1_lmo0444 (lmo0444), was
found in the Lm strain FDAARGOS_607, the Lm strain 4/52-1953, the Lm strain UKVDL7, the Lm strain NTSN and the Lm strain FSL-J1-158. Two genes of the SSI-2, associated in Lm with a
tolerance to alkaline and oxidative stresses, SSI2_lin0464 (lin0464) and SSI2_lin0465 (lin0465)112 and absent in the Lm strain AUF, were revealed in the Lm strain UKVDL4 and the Lm strain
UKVDL925. Importantly, we found pronounced polymorphisms in the majority of the alleles of the genes responsible for antibiotic resistance and motility, SSI-1 and determination of Lm Genomic
Islands in the Lm strain AUF compared with the other Lm strains used25. The Lm strain AUF demonstrated almost total identity exclusively with only the Lm strain EGD and the Lm strain 10403S
but not with other Lm strains independently from their genetic lineage, ST, and CC. In order to examine the specific contribution of UVR to the inactivation of the Lm strain AUF genes, we
compared all the 17 relevant pseudogenes found in the genome of this strain with other Lm genomes available in GenBank using the BLAST resource (https://blast.ncbi.nlm.nih.gov/Blast.cgi). We
identified only four putative genes, _No. 2_ (locus tag: GZH80_03675), _No. 4_ (locus tag: GZH80_06950), _No. 6_ (locus tag: GZH80_08750), and _No. 13_ (GZH80_11730), which were assigned in
the Lm strain AUF as pseudogenes, while in the majority of other Lm strains (up to 99-100 of 100 strains available) were annotated as genes encoding functional protein products113. These
proteins were the following: (i) 100 out 100 Lm strains: tetratricopeptide repeat protein, sensor histidine kinase; and amino acid permease, and (ii) 99 out of 100 Lm strains: a hypothetical
protein which was found as the pseudogene with 100% identity to the Lm strain AUF in only a single Lm strain 3453. The relevant genes demonstrated homology on the level of 92.87 – 99.93%
versus the Lm strain AUF as a result of the presence of either frameshift mutations in genes No. 2, 6, and 13, or missing C-terminus in the gene _No. 4_. The products expressed by these
genes are involved in bacterial pathogenesis by controlling the expression of virulence, biofilm formation, protein-protein interactions, antimicrobial resistance, quorum sensing and
signaling27,114,115,116. The relevant mutants in wild Lm strains have been reported to be defective for intracellular growth and cell-to-cell spread and were severely attenuated for
virulence in a mouse model114. At the same time, when the genome of the Lm strain AUF was compared with the genomes of its closest homolog, the Lm strain EGD, we found only a single
pseudogene (1/17 representation in the Lm strain AUF), _No. 9_ (LMON_0171), with possible expression of only a truncated hypothetical protein113. Furthermore, in the genome of the Lm strain
10403S four of 17 pseudogenes related to those in the Lm strain AUF78 due to frameshift mutations were annotated, such as: _No. 7_ (LMRG_02848), _No. 11_ (LMRG_00154), _No. 12_ (LMRG_00322)
and _No. 15_ (LMRG_02951). We believe some of these genes could be damaged by UVR resulting in the UVR-induced DNA mutations leading to the Lm strain AUF attenuation. USAGE NOTES Currently,
we present the complete genome sequence and the gene annotations for the Lm strain AUF, which has been used for decades as a live whole-cell veterinary vaccine. We compared the Lm strain AUF
genome with the whole genomes of reference fully virulent Lm strains, of the Lm isolates derived from animals with listeriosis, and the Lm strain used for the development of live attenuated
vaccine vectors against cancer. We believe that the data obtained will be useful to unravel the mechanisms of attenuation and virulence in _Listeria_ and other pathogenic microorganisms.
However, it is very difficult to make objective conclusions on the Lm strain AUF attenuation in the absence of the wild parental Lm strain ‘A’. Nevertheless, we hope that these data will be
valuable as the basic platform for development of the effective and safe new-generation vaccine(s) with improved characteristics for prophylaxis of listeriosis in animals worldwide . CODE
AVAILABILITY Software used in the generation or processing of our data is stated in the Methods section. Detailed information including versions of software and database are provided in
Table 5. REFERENCES * Seeliger, H. P. R. Listeriosis-history and actual developments. _Infection_ 16, S80–S84 (1988). Article PubMed Google Scholar * Lecuit, M. _Listeria monocytogenes_,
a model in infection biology. _Cellular microbiology_ 22, e13186 (2020). Article CAS PubMed Google Scholar * Farber, J. M. & Peterkin, P. _Listeria monocytogenes_, a food-borne
pathogen. _Microbiological reviews_ 55, 476–511 (1991). Article CAS PubMed PubMed Central Google Scholar * Dhama, K. _et al_. Listeriosis in animals, its public health significance
(food-borne zoonosis) and advances in diagnosis and control: a comprehensive review. _Vet Q_ 35, 211–235 (2015). Article MathSciNet PubMed Google Scholar * Koopmans, M. M., Brouwer, M.
C., Vázquez-Boland, J. A. & van de Beek, D. Human Listeriosis. _Clin Microbiol Rev_ 36, e00060192023 (2023). Article Google Scholar * Luo, X. & Cai, X. A combined use of autolysin
p60 and listeriolysin O antigens induces high protective immune responses against _Listeria monocytogenes_ infection. _Curr Microbiol_ 65, 813–8 (2012). Article CAS PubMed Google Scholar
* Félix, B. _et al_. A European-wide dataset to uncover adaptive traits of _Listeria monocytogenes_ to diverse ecological niches. _Sci Data_ 9, 190 (2022). Article PubMed PubMed Central
Google Scholar * Murray, E. G. D., Webb, R. E. & Swami, M. B. R. A disease of rabbits characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed _bacillus
Bacterium monocytogenes_ (_n.sp_.). _J Pathol Bacteriol_ 29, 407–439 (1926). Article Google Scholar * Williams, E. N., Van Doren, J. M., Leonard, C. L. & Datta, A. R. Prevalence of
_Listeria monocytogenes_, _Salmonella spp_., Shiga toxin-producing _Escherichia coli_, and _Campylobacter spp_. in raw milk in the United States between 2000 and 2019: A systematic review
and meta-analysis. _J Food Prot_ 86, 100014 (2023). Article PubMed Google Scholar * Esteban, J. I., Oporto, B., Aduriz, G., Juste, R. A. & Hurtado, A. Faecal shedding and strain
diversity of _Listeria monocytogenes_ in healthy ruminants and swine in Northern Spain. _BMC Vet Res_ 5, 2 (2009). Article PubMed PubMed Central Google Scholar * Gaulin, C., Ramsay, D.
& Bekal, S. Widespread listeriosis outbreak attributable to pasteurized cheese, which led to extensive cross-contamination affecting cheese retailers, Quebec, Canada, 2008. _J Food Prot_
75, 71–78 (2012). Article PubMed Google Scholar * Jackson, K. A. _et al_. Multistate outbreak of _Listeria monocytogenes_ associated with Mexican-style cheese made from pasteurized milk
among pregnant, Hispanic women. _J Food Prot_ 74, 949–953 (2011). Article CAS PubMed Google Scholar * Johnsen, B. O., Lingaas, E., Torfoss, D., Strom, E. H. & Nordoy, I. A large
outbreak of _Listeria monocytogenes_ infection with short incubation period in a tertiary care hospital. _J Infect_ 61, 465–470 (2010). Article PubMed Google Scholar * Altissimi, C.,
Noé-Nordberg, C., Ranucci, D. & Paulsen, P. Presence of Foodborne Bacteria in Wild Boar and Wild Boar Meat-A Literature Survey for the Period 2012-2022. _Foods_ 12, 1689 (2023). Article
PubMed PubMed Central Google Scholar * Thomas, J. _et al_. Outbreak of Listeriosis in South Africa Associated with Processed Meat. _N Engl J Med_ 382, 632–643 (2020). Article CAS
PubMed PubMed Central Google Scholar * Bevilacqua, A. _et al_. Microbiological Risk Assessment in Foods: Background and Tools, with a Focus on Risk Ranger. _Foods_ 12, 1483 (2023).
Article CAS PubMed PubMed Central Google Scholar * Ali, S. & Alsayeqh, A. F. Review of major meat-borne zoonotic bacterial pathogens. _Front Public Health_ 10, 1045599 (2022).
Article PubMed PubMed Central Google Scholar * Jibo, G. G. _et al_. A systematic review and meta-analysis of the prevalence of _Listeria monocytogenes_ in South-East Asia; a one-health
approach of human-animal-food-environment. _One Health_ 15, 100417 (2022). Article PubMed PubMed Central Google Scholar * Huang, C., Lu, T. L. & Yang, Y. Mortality risk factors
related to listeriosis - A meta-analysis. _J Infect Public Health_ 16, 771–783 (2023). Article PubMed Google Scholar * Osek, J. & Wieczorek, K. _Listeria monocytogenes_-How This
Pathogen Uses Its Virulence Mechanisms to Infect the Hosts. _Pathogens_ 11, 1491 (2022). Article CAS PubMed PubMed Central Google Scholar * Pogreba-Brown, K. _et al_. Complications
Associated with Foodborne Listeriosis: A Scoping Review. _Foodborne Pathog Dis_ 19, 725–743 (2022). Article CAS PubMed Google Scholar * Tan, W., Wang, G., Pan, Z., Yin, Y. & Jiao, X.
Complete Genome Sequence of _Listeria monocytogenes_ NTSN, a Serovar 4b and Animal Source Strain. _Genome Announc_ 3, e01403–14 (2015). Article PubMed PubMed Central Google Scholar *
Kichemazova, N. V., Khizhnyakova, M. A., Lyapina, A. M., Kolosova, A. A. & Feodorova, V. A. Vaccine prophylaxis of listeriosis in farm animals. _J. Veterinariya_ 3, 18–25 (2023). Google
Scholar * Selivanov, A. V., Kotylev, O. A. & Sedov, N. K. Listeriosis vaccine from strain AUF listeriae. _Veterinariia_ 12, 38–39 (1974). Google Scholar * Feodorova, V. _et al_.
Supplementary Table 1. _figshare_ https://doi.org/10.6084/m9.figshare.24298378.v6 (2024). * Moura, A. _et al_. Whole genome-based population biology and epidemiological surveillance of
_Listeria monocytogenes_. _Nature microbiology_ 2, 1–10 (2016). Article Google Scholar * Blatch, G. L. & Lässle, M. The tetratricopeptide repeat: a structural motif mediating
protein-protein interactions. _Bioessays_ 21, 932–9 (1999). Article CAS PubMed Google Scholar * Dussurget, O. New insights into determinants of _Listeria monocytogenes_ virulence. _Int
Rev Cell Mol Biol_ 270, 1–38 (2008). Article CAS PubMed Google Scholar * Gelbíčová, T., Kolácková, I., Pantucek, R. & Karpíšková, R. A novel mutation leading to a premature stop
codon in _inlA_ of _Listeria monocytogenes_ isolated from neonatal listeriosis. _New Microbiol_ 38, 293–296 (2015). PubMed Google Scholar * Maury, M. M. _et al_. Uncovering _Listeria
monocytogenes_ hypervirulence by harnessing its biodiversity. _Nat Genet_ 8, 308–313 (2016). Article Google Scholar * Quereda, J. J., Meza-Torres, J., Cossart, P. & Pizarro-Cerdá, J.
Listeriolysin S: A bacteriocin from epidemic _Listeria monocytogenes_ strains that targets the gut microbiota. _Gut Microbes_ 8, 384–391 (2017). Erratum for: Addendum to, Quereda, J. J. _et
al_. Bacteriocin from epidemic _Listeria_ strains alters the host intestinal microbiota to favor infection. _Proc Nat Acad Sci USA_ 113, 5706–11 (2016). Article CAS PubMed PubMed Central
Google Scholar * Lakicevic, B. Z., Den Besten, H. M. W. & De Biase, D. Landscape of Stress Response and Virulence Genes Among _Listeria monocytogenes_ Strains. _Front Microbiol_ 12,
738470 (2022). Article PubMed PubMed Central Google Scholar * Ryan, S., Begley, M., Hill, C. & Gahan, C. G. A five-gene stress survival islet (SSI-1) that contributes to the growth
of _Listeria monocytogenes_ in suboptimal conditions. _J Appl Microbiol_ 109, 984–995 (2010). Article CAS PubMed Google Scholar * Sichtig, H. _et al_. FDA-ARGOS is a database with public
quality-controlled reference genomes for diagnostic use and regulatory science. _Nat Commun_ 10, 3313 (2019). Article ADS PubMed PubMed Central Google Scholar * Database for Reference
Grade Microbial Sequences (FDA-ARGOS) https://www.fda.gov/medical-devices/science-and-research-medical-devices/database-reference-grade-microbial-sequences-fda-argos (2023). * Reference
genome ASM19603v1 European Consortium. Strain: EGD-e. https://www.ncbi.nlm.nih.gov/datasets/taxonomy/169963/ (2003). * Genome assembly ASM19603v1, reference
https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000196035.1/. * Bécavin, C. _et al_. Comparison of widely used _Listeria monocytogenes_ strains EGD, 10403S, and EGD-e highlights genomic
variations underlying differences in pathogenicity. _mBio_ 5, e00969–14 (2014). Article PubMed PubMed Central Google Scholar * Maciag, P. C., Radulovic, S. & Rothman, J. The first
clinical use of a live-attenuated _Listeria monocytogenes_ vaccine: A Phase I safety study of Lm-LLO-E7 in patients with advanced carcinoma of the cervix. _Vaccine_ 27, 3975–3983 (2009).
Article CAS PubMed Google Scholar * Flickinger, J. C. Jr., Rodeck, U. & Snook, A. E. _Listeria monocytogenes_ as a Vector for Cancer Immunotherapy: Current Understanding and
Progress. _Vaccines (Basel)_ 6, 48 (2018). Article CAS PubMed Google Scholar * Gunn, G. R. _et al_. Two _Listeria monocytogenes_ vaccine vectors that express different molecular forms of
human papilloma virus-16 (HPV-16) E7 induce qualitatively different T cell immunity that correlates with their ability to induce regression of established tumors immortalized by HPV-16. _J
Immunol_ 167, 6471–9 (2001). Article CAS PubMed Google Scholar * Gilley, R. P. & Dube, P. H. Checkpoint blockade inhibitors enhances the effectiveness of a _Listeria
monocytogenes_-based melanoma vaccine. _Oncotarget_ 11, 740–754 (2020). Article PubMed PubMed Central Google Scholar * Oladejo, M., Paterson, Y. & Wood, L. M. Clinical Experience and
Recent Advances in the Development of _Listeria_-Based Tumor Immunotherapies. _Front Immunol_ 12, 642316 (2021). Article CAS PubMed PubMed Central Google Scholar * Wood, L. M. &
Paterson, Y. Attenuated _Listeria monocytogenes_: a powerful and versatile vector for the future of tumor immunotherapy. _Front Cell Infect Microbiol_ 4, 51 (2014). Article PubMed PubMed
Central Google Scholar * Bespalova, T. Y. _et al_. Novel Sequence Types of _Listeria monocytogenes_ of Different Origin Obtained in the Republic of Serbia. _Microorganisms_ 9, 1289 (2021).
Article CAS PubMed PubMed Central Google Scholar * Zaitsev, S. S., Khizhnyakova, M. A. & Feodorova, V. A. Retrospective Investigation of the Whole Genome of the Hypovirulent
_Listeria monocytogenes_ Strain of ST201, CC69, Lineage III, Isolated from a Piglet with Fatal Neurolisteriosis. _Microorganisms_ 10, 1442 (2022). Article CAS PubMed PubMed Central
Google Scholar * Zaitsev, S., Khizhnyakova, M. A. & Feodorova, V. A. Data of _de novo_ genome assembly of the _Listeria monocytogenes_ vaccine strain AUF. _GenBank_
https://www.ncbi.nlm.nih.gov/nuccore/CP048400 (2020). * Li, W. _et al_. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. _Nucleic Acids
Res_ 49, D1020–D1028 (2021). Article ADS CAS PubMed Google Scholar * Bertels, F., Silander, O. K., Pachkov, M., Rainey, P. B. & van Nimwegen, E. Automated reconstruction of
whole-genome phylogenies from short-sequence reads. _Mol Biol Evol_ 31, 1077–88 (2014). Article CAS PubMed PubMed Central Google Scholar * Kumar, S., Stecher, G. & Tamura, K. MEGA7:
Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. _Mol Biol Evol_ 33, 1870–4 (2016). Article CAS PubMed PubMed Central Google Scholar * Olson, R. D. _et al_.
Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. _Nucleic Acids Res_ 51, D678–D689 (2023). Article CAS PubMed Google
Scholar * Grant, J. R. _et al_. Proksee: in-depth characterization and visualization of bacterial genomes Nucleic Acids Research, gkad326, https://doi.org/10.1093/nar/gkad326 2023. *
Mason, C. _et al_. FDA dAtabase for Regulatory Grade micrObial Sequences (FDA-ARGOS): Supporting development and validation of Infectious Disease Dx tests. _GenBank_
https://www.ncbi.nlm.nih.gov/nuccore/CP041014.1 (2019). * Glaser, P. _et al_. Comparative genomics of _Listeria_ species. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/AL591824 (2001). *
Becavin, C. _et al_. Comparison of widely used _Listeria monocytogenes_ strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. _GenBank_
https://www.ncbi.nlm.nih.gov/nuccore/HG421741.1 (2014). * Bechtel, T. & Gibbons, J. Genome sequence of three persistent _L. monocytogenes_ strains isolated from a food processing
facility and a livestock outbreak. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/CP090057.1 (2022). * Tan, W., Wang, G., Pan, Z., Yin, Y. & Jiao, X. Complete Genome Sequence of
_Listeria monocytogenes_ NTSN, a Serovar 4b and Animal Source Strain. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/CP009897.1 (2015). * Senay, T. E. _et al_. Neurotropic Lineage III
Strains of _Listeria monocytogenes_ Disseminate to the Brain without Reaching High Titer in the Blood. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/CP065028.1 (2020). * Senay, T. E. _et
al_. Neurotropic Lineage III Strains of _Listeria monocytogenes_ Disseminate to the Brain without Reaching High Titer in the Blood. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/CP076644.1
(2021). * Zaitsev, S., Khizhnyakova, M. A., Filonova, N. & Feodorova, V. A. Data of _de novo_ genome assembly of the _Listeria monocytogenes_ strain of zoonotic origin. _GenBank_
https://www.ncbi.nlm.nih.gov/nuccore/CP048401.1 (2020). * Senay, T. E. _et al_. Neurotropic Lineage III Strains of _Listeria monocytogenes_ Disseminate to the Brain without Reaching High
Titer in the Blood. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/CP076669.1 (2020). * Borowsky, M. _et al_. The Genome Sequence of _Listeria monocytogenes_ strain 10403S. _GenBank_
https://www.ncbi.nlm.nih.gov/nuccore/NC_017544.1 (2010). * Wellcome Trust Sanger Institute. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/LR134483.1 (2018). * Quereda, J. J. _et al_.
_Listeria valentina_ sp. nov., isolated from a water trough and the faeces of healthy sheep. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/JAATJW010000100 (2020). * Schardt, J.,
Mueller-Herbst, S., Scherer, S. & Huptas, C. Direct Submission. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/LARY01000001 (2015). * Den Bakker, H. C. _et al_. _Listeria floridensis_
sp. nov., _Listeria aquatica_ sp. nov., _Listeria cornellensis_ sp. nov., _Listeria riparia_ sp. nov. and _Listeria grandensis_ sp. nov., from agricultural and natural environments.
_GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/NZ_AODF00000000 (2014). * Chiara, M. _et al_. Draft genome sequences of _L. newyorkensis_ and _L. fleischmannii_ clarify phylogenetic
relationships within the genus _Listeria_ and provide additional insights into the evolution of pathogenicity in _Listeria sensu strictu_. _GenBank_
https://www.ncbi.nlm.nih.gov/nuccore/KQ130608 (2015). * Weller, D. _et al_. _Listeria ohnekaius_ sp. nov. and _Listeria portnoyii_ sp. nov. isolated from non-agricultural and natural
environments. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/NZ_JABJVM000000000.1 (2023). * Bell, G., Cornelius, A. & Taylor, W. _Listeria newyorkensis_ isolated from tuaki (NZ cockles).
_GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP113980.1 (2023). * Den Bakker, H. C. _et al_. _Listeria floridensis_ sp. nov., _Listeria aquatica_ sp. nov., _Listeria cornellensis_ sp.
nov., _Listeria riparia_ sp. nov. and _Listeria grandensis_ sp. nov., from agricultural and natural environments. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/NZ_AODE00000000.1 (2014). *
Whitman, W. Genomic Encyclopedia of Type Strains, Phase III (KMG-III): the genomes of soil and plant-associated and newly described type strains. _GenBank_
https://www.ncbi.nlm.nih.gov/nuccore/NZ_SNZK01000001 (2019). * Weller, D., Andrus, A., Wiedmann, M. & den Bakker, H. C. _Listeria booriae_ sp. nov. and _Listeria newyorkensis_ sp. nov.,
from food processing environments in the USA. _GenBank_ https://www.ncbi.nlm.nih.gov/nuccore/NZ_JNFA00000000.1 (2015). * Loessner, M. J., Inman, R. B., Lauer, P. & Calendar, R. Complete
nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of _Listeria monocytogenes_: implications for phage evolution. _GenBank_
https://www.ncbi.nlm.nih.gov/nuccore/NC_003216.1 (2000). * _NCBI Sequence Read Archive_ https://identifiers.org/ncbi/insdc.sra:SRR25210281 (2023). * _NCBI Sequence Read Archive_
https://identifiers.org/ncbi/insdc.sra:SRR25180708 (2023). * Feodorova, V. _et al_. Supplementary Table 2. _figshare_ https://doi.org/10.6084/m9.figshare.24299374.v1 (2023). *
Espinoza-Mellado, M. D. R. & Vilchis-Rangel, R. E. Review of CRISPR-Cas Systems in _Listeria_ Species: Current Knowledge and Perspectives. _Int J Microbiol_ 2022, 9829770 (2022). Article
PubMed PubMed Central Google Scholar * Mihara, T. _et al_. Linking virus genomes with host taxonomy. _Viruses_ 8, 66 (2016). Article PubMed PubMed Central Google Scholar * Loessner,
M. J., Inman, R. B., Lauer, P. & Calendar, R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of _Listeria monocytogenes_: implications for
phage evolution. _Mol Microbiol_ 35, 324–40 (2000). Article CAS PubMed Google Scholar * Loessner, M. J., Wendlinger, G. & Scherer, S. Heterogeneous endolysins in _Listeria
monocytogenes_ bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. _Mol Microbiol_ 16, 1231–41 (1995). Article CAS PubMed
Google Scholar * Hu, M. _et al_. A Meta-Analysis and Systematic Review of _Listeria monocytogenes_ Response to Sanitizer Treatments. _Foods_ 12, 154 (2022). Article PubMed PubMed Central
Google Scholar * McCafferty, D. G. & Melvin, J. A. _Handbook of Proteolytic Enzymes_ (Academic Press is an imprint of Elsevier, 2013). * Klebba, P. E., Charbit, A., Xiao, Q., Jiang,
X. & Newton, S. M. Mechanisms of iron and haem transport by _Listeria monocytogenes_. _Mol Membr Biol_ 29, 69–86 (2012). Article CAS PubMed Google Scholar * Gray, B., Hall, P. &
Gresham, H. Targeting _agr_- and _agr_-Like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. _Sensors (Basel)_ 13, 5130–66
(2013). Article ADS CAS PubMed Google Scholar * Autret, N., Raynaud, C., Dubail, I., Charbit, A. & Berche, P. Identification of the _agr_ locus of _Listeria monocytogenes_: Role in
bacterial virulence. _Infec Immun_ 71, 4463–4471 (2003). Article CAS Google Scholar * Rieu, A., Weidmann, S., Garmyn, D., Piveteau, P. & Guzzo, J. Agr system of _Listeria
monocytogenes_ EGD-e: Role in adherence and differential expression pattern. _Appl Environ Microbiol_ 73, 6125–6133 (2007). Article ADS CAS PubMed PubMed Central Google Scholar *
Riedel, C. U. _et al_. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in _Listeria monocytogenes_. _Mol Microbiol_ 71,
1177–1189 (2009). Article CAS PubMed Google Scholar * Gupta, V.K. & Jindal, V. in _Integrated Pest Management Current Concepts and Ecological Perspective_ (ed. Abrol D. P.) Ch. 16
(Elsevier, 2014). * Vornhagen, J. _et al_. The _Klebsiella pneumoniae_ citrate synthase gene, _gltA_, influences site specific fitness during infection. _PLoS Pathog_ 15, e1008010 (2019).
Article CAS PubMed PubMed Central Google Scholar * Cabanes, D., Dussurget, O., Dehoux, P. & Cossart, P. Auto, a surface associated autolysin of _Listeria monocytogenes_ required for
entry into eukaryotic cells and virulence. _Mol Microbiol_ 51, 1601–1614 (2004). Article CAS PubMed Google Scholar * Wang, L. & Lin, M. A novel cell wall-anchored peptidoglycan
hydrolase (autolysin), IspC, essential for _Listeria monocytogenes_ virulence: Genetic and proteomic analysis. _Microbiol_ 154, 1900–1913 (2008). Article CAS Google Scholar *
Leimeister-Wächter, M., Haffner, C., Domann, E., Goebel, W. & Chakraborty, T. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor
of _Listeria monocytogenes_. _Proc Natl Acad Sci USA_ 87, 8336–8340 (1990). Article ADS PubMed PubMed Central Google Scholar * Mengaud, J. _et al_. Pleiotropic control of _Listeria
monocytogenes_ virulence factors by a gene that is autoregulated. _Mol Microbiol_ 5, 2273–2283 (1991). Article CAS PubMed Google Scholar * De las Heras, A., Cain, R. J., Bielecka, M. K.
& Vázquez-Boland, J. A. Regulation of _Listeria_ virulence: PrfA master and commander. _Curr Opin Microbiol_ 14, 118–127 (2011). Article Google Scholar * Feodorova, V. _et al_.
Supplementary Table 4. _figshare_ https://doi.org/10.6084/m9.figshare.24298378.v1 (2023). * Feodorova, V. _et al_. Alignment of the nucleotide (A) and amino acid (B) sequences of the _prfA_
gene derived from the _Listeria monocytogenes_ strains AUF, EGD-e, EGD, NTSN, UKVDL4, UKVDL9, UKVDL7, 4/52-1953, FDAARGOS 607 and FSL-J1-158, respectively. _figshare_
https://doi.org/10.6084/m9.figshare.24796788.v5 (2024). * Portnoy, D. A., Jacks, P. S. & Hinrichs, D. J. Role of hemolysin for the intracellular growth of _Listeria monocytogenes_. _J
Exp Med_ 167, 1459–1471 (1988). Article CAS PubMed Google Scholar * Feodorova, V. _et al_. Alignment of the nucleotide (A) and amino acid (B) sequences of the _hly_ gene derived from the
_Listeria monocytogenes_ strains AUF, EGD, EGD-e, FDAARGOS 607, FSL-J1-158, UKVDL7 and 4/52-1953. _figshare_ https://doi.org/10.6084/m9.figshare.24796863.v6 (2024). * Ireton, K., Mortuza,
R., Gyanwali, G. C., Gianfelice, A. & Hussain, M. Role of internalin proteins in the pathogenesis of _Listeria monocytogenes_. _Mol Microbiol_ 116, 1407–1419 (2021). Article CAS PubMed
Google Scholar * Tsai, Y. H., Orsi, R. H., Nightingale, K. K. & Wiedmann, M. _Listeria monocytogenes_ internalins are highly diverse and evolved by recombination and positive
selection. _Infect Genet Evol_ 6, 378–89 (2006). Article CAS PubMed Google Scholar * Rychli, K. _et al_. _Listeria monocytogenes_ Isolated from Illegally Imported Food Products into the
European Union Harbor Different Virulence Factor Variants. _Genes (Basel)_ 9, 428 (2018). Article PubMed Google Scholar * Feodorova, V. _et al_. Alignment of the nucleotide (A) and amino
acid (B) sequences of the _inlA_ gene derived from the _Listeria monocytogenes_ strains AUF, EGD, EGD-e, FDAARGOS 607, NTSN, UKVDL4, 4/52-1953 and UKVDL7. _figshare_
https://doi.org/10.6084/m9.figshare.24796872.v4 (2024). * Feodorova, V. _et al_. Alignment of the nucleotide (A) and amino acid (B) sequences of the _inlB_ gene derived from the _Listeria
monocytogenes_ strains AUF, EGD, EGD-e, FDAARGOS 607, NTSN, UKVDL4, 4/52-1953 and FSL-J1-158. _figshare_ https://doi.org/10.6084/m9.figshare.24796878.v4 (2024). * Raffelsbauer, D. _et al_.
The gene cluster inlC2DE of _Listeria monocytogenes_ contains additional new internalin genes and is important for virulence in mice. _Mol Gen Genet_ 260, 144–58 (1998). Article CAS PubMed
Google Scholar * Fillgrove, K. L., Pakhomova, S., Schaab, M. R., Newcomer, M. E. & Armstrong, R. N. Structure and mechanism of the genomically encoded fosfomycin resistance protein,
FosX, from _Listeria monocytogenes_. _Biochemistry_ 46, 8110–8120 (2007). Article CAS PubMed Google Scholar * Thedieck, K. _et al_. The MprF protein is required for lysinylation of
phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on _Listeria monocytogenes_. _Mol Microbiol_ 62, 1325–1339 (2006). Article CAS PubMed
Google Scholar * Dar, D. _et al_. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. _Science_ 352, aad9822 (2016). Article PubMed PubMed Central Google
Scholar * Haubert, L., Kremer, F. S. & da Silva, W. P. Whole-genome sequencing identification of a multidrug-resistant _Listeria monocytogenes_ serotype 1/2a isolated from fresh mixed
sausage in southern Brazil. _Infect Genet Evol_ 65, 127–130 (2018). Article CAS PubMed Google Scholar * Lebrun, M., Audurier, A. & Cossart, P. Plasmid-borne cadmium resistance genes
in _Listeria monocytogenes_ are similar to _cadA_ and _cadC_ of _Staphylococcus aureus_ and are induced by cadmium. _J Bacteriol_ 176, 3040–3048 (1994). Article CAS PubMed PubMed Central
Google Scholar * Parsons, C., Lee, S. & Kathariou, S. Heavy Metal Resistance Determinants of the Foodborne Pathogen _Listeria monocytogenes_. _Genes (Basel)_ 24, 11 (2018). Article
Google Scholar * Harter, E. _et al_. Stress Survival Islet 2, Predominantly Present in _Listeria Monocytogenes_ Strains of Sequence Type 121, Is Involved in the Alkaline and Oxidative
Stress Responses. _Appl Environ Microbiol_ 83, e00827–17 (2017). Article CAS PubMed PubMed Central Google Scholar * Feodorova, V. _et al_. Supplementary Table 3. _figshare_
https://doi.org/10.6084/m9.figshare.24299377.v4 (2024). * Sun, A. N., Camilli, A. & Portnoy, D. A. Isolation of _Listeria monocytogenes_ small-plaquemutants defective for intracellular
growth and cell-to-cell spread. _Infect Immun_ 58, 3770–3778 (1990). Article CAS PubMed PubMed Central Google Scholar * Vaval, T. D. M., Xayarath, B. & Freitag, N. E. Two Permeases
Associated with the Multifunctional CtaP Cysteine Transport System in _Listeria monocytogenes_ Play Distinct Roles in Pathogenesis. _Microbiol Spectr_ 11, e0331722 (2023). Article Google
Scholar * Ishii, E. & Eguchi, Y. Diversity in Sensing and Signaling of Bacterial Sensor Histidine Kinases. _Biomolecules_ 11, 1524 (2021). Article CAS PubMed PubMed Central Google
Scholar * Glaser, P. _et al_. Comparative genomics of _Listeria_ species. _Science_ 294, 849–52 (2001). Article ADS CAS PubMed Google Scholar * Toledo-Arana, A. _et al_. The _Listeria_
transcriptional landscape from saprophytism to virulence. _Nature_ 459, 950–6 (2009). Article ADS CAS PubMed Google Scholar * Chatterjee, S. S. _et al_. Intracellular gene expression
profile of _Listeria monocytogenes_. _Infect Immun_ 74, 1323–38 (2006). Article CAS PubMed PubMed Central Google Scholar * Den Bakker, H., Bowen, B., Rodriguez-Rivera, L. &
Wiedmann, M. FSL J1-208, a Virulent Uncommon Phylogenetic Lineage IV _Listeria monocytogenes_ Strain with a Small Chromosome Size and a Putative Virulence Plasmid Carrying Internalin-Like
Genes. _Applied and environmental microbiology_ 78, 1876–89 (2012). Article ADS Google Scholar * Senay, T. E. _et al_. Neurotropic Lineage III Strains of _Listeria monocytogenes_
Disseminate to the Brain without Reaching High Titer in the Blood. _mSphere_ 5, e00871–20 (2020). Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS
The reported study was funded by Russian Science Foundation, the project number 22-16-00165. We would like to thank Dr. Daria G. Oglodina for her technical assistance in the manuscript
preparation. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Laboratory for Fundamental and Applied Research, Saratov State University of Genetics, Biotechnology and Engineering named after
N.I. Vavilov, Saratov, Russia Valentina A. Feodorova, Sergey S. Zaitsev, Mariya A. Khizhnyakova, Maxim S. Lavrukhin, Yury V. Saltykov & Olga S. Larionova * Department for Microbiology
and Biotechnology, Saratov State University of Genetics, Biotechnology and Engineering named after N.I. Vavilov, Saratov, Russia Valentina A. Feodorova & Olga S. Larionova * All-Russian
Scientific Research and Technological Institute of Biological Industry, Biocombinat, Moscow, Russia Alexey D. Zaberezhny Authors * Valentina A. Feodorova View author publications You can
also search for this author inPubMed Google Scholar * Sergey S. Zaitsev View author publications You can also search for this author inPubMed Google Scholar * Mariya A. Khizhnyakova View
author publications You can also search for this author inPubMed Google Scholar * Maxim S. Lavrukhin View author publications You can also search for this author inPubMed Google Scholar *
Yury V. Saltykov View author publications You can also search for this author inPubMed Google Scholar * Alexey D. Zaberezhny View author publications You can also search for this author
inPubMed Google Scholar * Olga S. Larionova View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS V.A.F. provided funding, designed and
supervised the study, analysed the data, and wrote the manuscript, M.A.K. optimized and performed the sequencing, S.S.Z., and M.S.L. performed the bioinformatic analyses and prepared
graphical visualization of the data obtained, Y.V.S. contributed in computational analysis, A.D.Z., and O.S.L. provided material support, and edited the manuscript. CORRESPONDING AUTHOR
Correspondence to Valentina A. Feodorova. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TABLE 1 SUPPLEMENTARY TABLE 2 SUPPLEMENTARY TABLE 3
SUPPLEMENTARY TABLE 4 RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation,
distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and
indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to
the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will
need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Feodorova, V.A., Zaitsev, S.S., Khizhnyakova, M.A. _et al._ Complete genome of the _Listeria monocytogenes_ strain AUF, used as a live listeriosis veterinary vaccine. _Sci
Data_ 11, 643 (2024). https://doi.org/10.1038/s41597-024-03440-8 Download citation * Received: 08 August 2023 * Accepted: 28 May 2024 * Published: 17 June 2024 * DOI:
https://doi.org/10.1038/s41597-024-03440-8 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative