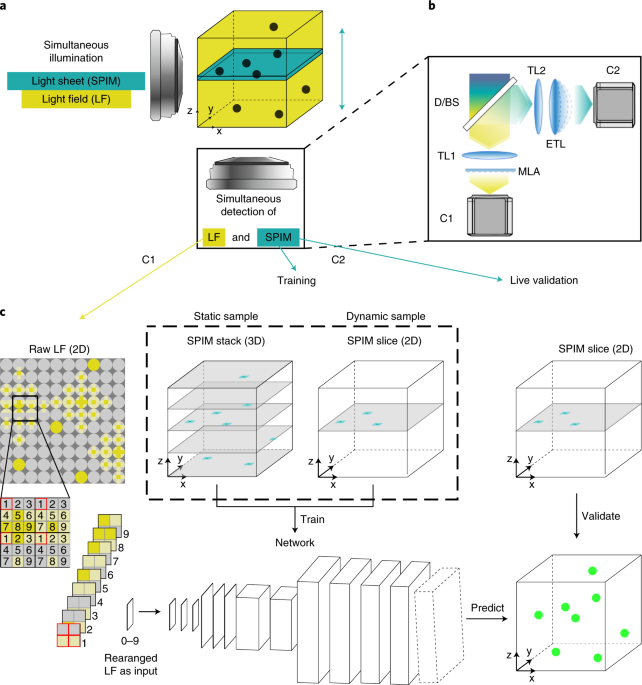
Deep learning-enhanced light-field imaging with continuous validation
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Visualizing dynamic processes over large, three-dimensional fields of view at high speed is essential for many applications in the life sciences. Light-field microscopy (LFM) has
emerged as a tool for fast volumetric image acquisition, but its effective throughput and widespread use in biology has been hampered by a computationally demanding and artifact-prone image
reconstruction process. Here, we present a framework for artificial intelligence–enhanced microscopy, integrating a hybrid light-field light-sheet microscope and deep learning–based volume
reconstruction. In our approach, concomitantly acquired, high-resolution two-dimensional light-sheet images continuously serve as training data and validation for the convolutional neural
network reconstructing the raw LFM data during extended volumetric time-lapse imaging experiments. Our network delivers high-quality three-dimensional reconstructions at video-rate
throughput, which can be further refined based on the high-resolution light-sheet images. We demonstrate the capabilities of our approach by imaging medaka heart dynamics and zebrafish
neural activity with volumetric imaging rates up to 100 Hz. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS
OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn
more Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to
full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our
FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS REAL-TIME VOLUMETRIC RECONSTRUCTION OF BIOLOGICAL DYNAMICS WITH LIGHT-FIELD MICROSCOPY AND DEEP LEARNING Article 11
February 2021 VIRTUAL-SCANNING LIGHT-FIELD MICROSCOPY FOR ROBUST SNAPSHOT HIGH-RESOLUTION VOLUMETRIC IMAGING Article Open access 06 April 2023 VOLUMETRIC IMAGING OF FAST CELLULAR DYNAMICS
WITH DEEP LEARNING ENHANCED BIOLUMINESCENCE MICROSCOPY Article Open access 03 December 2022 DATA AVAILABILITY The datasets generated and/or analyzed during the current study are available at
https://doi.org/10.5281/zenodo.4020352, https://doi.org/10.5281/zenodo.4020404 and https://doi.org/10.5281/zenodo.4019246. Links to additional datasets are provided at
https://github.com/kreshuklab/hylfm-net. Source data are provided with this paper. CODE AVAILABILITY The neural network code with routines for training and inference are available at
https://doi.org/10.5281/zenodo.4647764. REFERENCES * Winter, P. W. & Shroff, H. Faster fluorescence microscopy: advances in high speed biological imaging. _Curr. Opin. Chem. Biol._ 20,
46–53 (2014). Article CAS Google Scholar * Katona, G. et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. _Nat. Methods_ 9,
201–208 (2012). Article CAS Google Scholar * Duemani, R. G., Kelleher, K., Fink, R. & Saggau, P. Three-dimensional random access multiphoton microscopy for functional imaging of
neuronal activity. _Nat. Neurosci._ 11, 713–720 (2008). Article Google Scholar * Kong, L. et al. Continuous volumetric imaging via an optical phase-locked ultrasound lens. _Nat. Methods_
12, 759–762 (2015). Article CAS Google Scholar * Kazemipour, A. et al. Kilohertz frame-rate two-photon tomography. _Nat. Methods_ 16, 778–786 (2019). Article CAS Google Scholar *
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E. H. K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. _Science_ 305, 1007–1009
(2004). Article CAS Google Scholar * Bouchard, M. B. et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. _Nat.
Photonics_ 9, 113–119 (2015). Article CAS Google Scholar * Lu, R. et al. Video-rate volumetric functional imaging of the brain at synaptic resolution. _Nat. Neurosci._ 20, 620–628 (2017).
Article CAS Google Scholar * Quirin, S., Peterka, D. S. & Yuste, R. Instantaneous three-dimensional sensing using spatial light modulator illumination with extended depth of field
imaging. _Opt. Express_ 21, 16007–16021 (2013). Article Google Scholar * Levoy, M., Ng, R., Adams, A., Footer, M. & Horowitz, M. Light field microscopy. _ACM Trans. Graph._ 25, 924
(2006). Article Google Scholar * Broxton, M. et al. Wave optics theory and 3-D deconvolution for the light field microscope. _Opt. Express_ 21, 25418 (2013). Article Google Scholar *
Prevedel, R. et al. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. _Nat. Methods_ 11, 727–730 (2014). Article CAS Google Scholar * Aimon, S. et
al. Fast near-whole–brain imaging in adult _Drosophila_ during responses to stimuli and behavior. _PLoS Biol._ 17, e2006732 (2019). Article Google Scholar * Nöbauer, T. et al. Video rate
volumetric Ca2+ imaging across cortex using seeded iterative demixing (SID) microscopy. _Nat. Methods_ 14, 811–818 (2017). Article Google Scholar * Lin, Q. et al. Cerebellar neurodynamics
predict decision timing and outcome on the single-trial level. _Cell_ 180, 536–551 (2020). Article CAS Google Scholar * Wagner, N. et al. Instantaneous isotropic volumetric imaging of
fast biological processes. _Nat. Methods_ 16, 497–500 (2019). Article CAS Google Scholar * Truong, T. V. et al. High-contrast, synchronous volumetric imaging with selective volume
illumination microscopy. _Commun. Biol._ 3, 74 (2020). Article Google Scholar * Taylor, M. A., Nöbauer, T., Pernia-Andrade, A., Schlumm, F. & Vaziri, A. Brain-wide 3D light-field
imaging of neuronal activity with speckle-enhanced resolution. _Optica_ 5, 345 (2018). Article Google Scholar * Cohen, N. et al. Enhancing the performance of the light field microscope
using wavefront coding. _Opt. Express_ 22, 24817–24839 (2014). Article Google Scholar * Li, H. et al. Fast, volumetric live-cell imaging using high-resolution light-field microscopy.
_Biomed. Opt. Express_ 10, 29 (2019). Article CAS Google Scholar * Stefanoiu, A., Page, J., Symvoulidis, P., Westmeyer, G. G. & Lasser, T. Artifact-free deconvolution in light field
microscopy. _Opt. Express_ 27, 31644 (2019). Article Google Scholar * Scrofani, G. et al. FIMic: design for ultimate 3D-integral microscopy of in-vivo biological samples. _Biomed. Opt.
Express_ 9, 335 (2018). Article CAS Google Scholar * Guo, C., Liu, W., Hua, X., Li, H. & Jia, S. Fourier light-field microscopy. _Opt. Express_ 27, 25573 (2019). Article Google
Scholar * Guo, M. et al. Rapid image deconvolution and multiview fusion for optical microscopy. _Nat. Biotechnol._ 38, 1337–1346 (2020). * Schuler, C. J., Hirsch, M., Harmeling, S. &
Scholkopf, B. Learning to Deblur. _IEEE Trans. Pattern Anal. Mach. Intell._ 38, 1439–1451 (2016). Article Google Scholar * Belthangady, C. & Royer, L. A. Applications, promises, and
pitfalls of deep learning for fluorescence image reconstruction. _Nat. Methods_ 16, 1215–1225 (2019). Article CAS Google Scholar * Weigert, M. et al. Content-aware image restoration:
pushing the limits of fluorescence microscopy. _Nat. Methods_ 15, 1090–1097 (2018). Article CAS Google Scholar * Hoffman, D. P., Slavitt, I. & Fitzpatrick, C. A. The promise and peril
of deep learning in microscopy. _Nat. Methods_ 18, 131–132 (2021). Article CAS Google Scholar * Page, J., Saltarin, F., Belyaev, Y., Lyck, R. & Favaro, P. Learning to reconstruct
confocal microscopy stacks from single light field images. Preprint at https://arxiv.org/abs/2003.11004 (2020). * Wang, Z. et al. Real-time volumetric reconstruction of biological dynamics
with light-field microscopy and deep learning. _Nat. Methods_ https://doi.org/10.1038/s41592-021-01058-x (2021). * Fahrbach, F. O., Voigt, F. F., Schmid, B., Helmchen, F. & Huisken, J.
Rapid 3D light-sheet microscopy with a tunable lens. _Opt. Express_ 21, 21010 (2013). Article Google Scholar * Li, X. et al. DeepLFM: deep learning-based 3D reconstruction for light field
microscopy. In _Proc. Biophotonics Congress: Optics in the Life Sciences Congress 2019_ (BODA,BRAIN,NTM,OMA,OMP) NM3C.2 (Optical Society of America, 2019);
https://doi.org/10.1364/NTM.2019.NM3C.2 * Krull, A., Buchholz, T.-O. & Jug, F. Noise2Void—learning denoising from single noisy images. In _Proc. 2019 IEEE/CVF Conference on Computer
Vision and Pattern Recognition (CVPR)_ 2124–2132 (IEEE, 2019); https://doi.org/10.1109/CVPR.2019.00223 * Kobayashi, H., Solak, A. C., Batson, J. & Royer, L. A. Image deconvolution via
noise-tolerant self-supervised inversion. Preprint at https://arxiv.org/abs/2006.06156 (2020). * Preibisch, S. et al. Efficient Bayesian-based multiview deconvolution. _Nat. Methods_ 11,
645–648 (2014). Article CAS Google Scholar * Biewald, L. Experiment tracking with Weights and Biases. https://www.wandb.com/ (2021). * Shivanandan, A., Radenovic, A. & Sbalzarini, I.
F. MosaicIA: an ImageJ/Fiji plugin for spatial pattern and interaction analysis. _BMC Bioinf._ 14, 349 (2013). Article Google Scholar * Wang, Z., Simoncelli, E. P. & Bovik, A. C.
Multiscale structural similarity for image quality assessment. In _Proceedings of the_ _Thirty-Seventh Asilomar Conference on Signals, Systems & Computers, 2003_ 1398–1402 (IEEE, 2003);
https://doi.org/10.1109/ACSSC.2003.1292216 * Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. _Nat. Methods_ 9, 676–682 (2012). Article CAS Google Scholar
* Koster, R., Stick, R., Loosli, F. & Wittbrodt, J. Medaka spalt acts as a target gene of hedgehog signaling. _Development_ 124, 3147–3156 (1997). Article CAS Google Scholar *
Rembold, M., Lahiri, K., Foulkes, N. S. & Wittbrodt, J. Transgenesis in fish: efficient selection of transgenic fish by co-injection with a fluorescent reporter construct. _Nat. Protoc._
1, 1133–1139 (2006). Article CAS Google Scholar * Pnevmatikakis, E. A. & Giovannucci, A. NoRMCorre: an online algorithm for piecewise rigid motion correction of calcium imaging data.
_J. Neurosci. Methods_ 291, 83–94 (2017). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank the European Molecular Biology Laboratory (EMBL) Heidelberg mechanical
and electronic workshop for help as well as the IT Services Department for high-performance computing cluster support and C. Tischer from CBA for his help with volume registration. We thank
B. Balázs, Luxendo GmbH, for help with software and electronics, R. Singh and D. Kromm for general support and C. Pape for help with benchmarking the algorithms. We also thank M. Majewsky,
E. Leist and A. Saraceno for fish husbandry, and K. Slanchev and H. Baier (MPI Martinsried) as well as M. Hoffmann and B. Judkewitz (Charite Berlin) for providing calcium reporter zebrafish
lines. N.W. was supported by the Helmholtz Association under the joint research school Munich School for Data Science (MUDS). J.G. was supported by a Research Center for Molecular Medicine
(HRCMM) Career Development Fellowship, the MD/PhD program of the Medical Faculty Heidelberg, the Deutsche Herzstiftung e.V. (S/02/17), and by an Add-On Fellowship for Interdisciplinary
Science of the Joachim Herz Stiftung and is grateful to M. Gorenflo for supervision and guidance. N.N acknowledges support from Åke Wiberg foundation, Ingabritt and Arne Lundberg foundation,
and Sten K Johnson Foundation. J.C.B. acknowledges supporting fellowships from the EMBL Interdisciplinary Postdoctoral Programme under Marie Skłodowska Curie Cofund Actions MSCACOFUND-FP
(664726). L.H. thanks Luxendo GmbH for help with microscope software and equipment support. This work was supported by the European Molecular Biology Laboratory (F.B., N.W., N.N., J.C.B.,
L.H., A.K. and R.P.). AUTHOR INFORMATION Author notes * Nils Wagner Present address: Department of Informatics, Technical University of Munich, Garching, Germany * Nils Wagner Present
address: Munich School for Data Science (MUDS), Munich, Germany * These authors contributed equally: Nils Wagner, Fynn Beuttenmueller, Robert Prevedel and Anna Kreshuk. AUTHORS AND
AFFILIATIONS * Cell Biology and Biophysics Unit, European Molecular Biology Laboratory, Heidelberg, Germany Nils Wagner, Fynn Beuttenmueller, Nils Norlin, Juan Carlos Boffi, Lars Hufnagel,
Robert Prevedel & Anna Kreshuk * Collaboration for joint PhD degree between EMBL and Heidelberg University, Faculty of Biosciences, Heidelberg University, Heidelberg, Germany Fynn
Beuttenmueller * Department of Experimental Medical Science, Lund University, Lund, Sweden Nils Norlin * Lund Bioimaging Centre, Lund University, Lund, Sweden Nils Norlin * Centre for
Organismal Studies, Heidelberg University, Heidelberg, Germany Jakob Gierten & Joachim Wittbrodt * Department of Pediatric Cardiology, University Hospital Heidelberg, Heidelberg, Germany
Jakob Gierten * Institute of Bioengineering, School of Life Sciences, EPFL, Lausanne, Switzerland Martin Weigert * Developmental Biology Unit, European Molecular Biology Laboratory,
Heidelberg, Germany Robert Prevedel * Epigenetics and Neurobiology Unit, European Molecular Biology Laboratory, Monterotondo, Italy Robert Prevedel * Molecular Medicine Partnership Unit
(MMPU), European Molecular Biology Laboratory, Heidelberg, Germany Robert Prevedel Authors * Nils Wagner View author publications You can also search for this author inPubMed Google Scholar
* Fynn Beuttenmueller View author publications You can also search for this author inPubMed Google Scholar * Nils Norlin View author publications You can also search for this author inPubMed
Google Scholar * Jakob Gierten View author publications You can also search for this author inPubMed Google Scholar * Juan Carlos Boffi View author publications You can also search for this
author inPubMed Google Scholar * Joachim Wittbrodt View author publications You can also search for this author inPubMed Google Scholar * Martin Weigert View author publications You can
also search for this author inPubMed Google Scholar * Lars Hufnagel View author publications You can also search for this author inPubMed Google Scholar * Robert Prevedel View author
publications You can also search for this author inPubMed Google Scholar * Anna Kreshuk View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS
A.K., L.H. and R.P. conceived the project. N.W. and N.N. built the imaging system and performed experiments with the help of J.G. J.G. generated transgenic animals under guidance of J.W.
F.B., A.K. and M.W. conceived the CNN architecture. F.B. and N.W. implemented the CNN and other image processing parts of the computational pipeline and evaluated its performance. J.C.B.
performed Ca2+ data analysis. A.K. and R.P. led the project and wrote the paper with input from all authors. Author order for equal contributions was determined by coin toss. CORRESPONDING
AUTHORS Correspondence to Robert Prevedel or Anna Kreshuk. ETHICS DECLARATIONS COMPETING INTERESTS L.H. is scientific cofounder and employee of Luxendo GmbH (part of Bruker), which makes
light-sheet-based microscopes commercially available, and a shareholder of Suricube GmbH, which makes high-performance electronics and software for instrumentation control available. The
other authors declare no competing interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature Methods_ thanks the anonymous reviewers for their contribution to the peer review of this
work. Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. PUBLISHER’S NOTE Springer
Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 NETWORK ARCHITECTURE. This architecture was
used for beads and neural activity volumes. For the medaka heart, slightly different layer depth was used with the same overall structure (see Supplementary Table 1). Res2/3d: residual
blocks with 2d or 3d convolutions with kernel size (3×)3 × 3. Residual blocks contain an additional projection layer (1 × 1 or 1 × 1 × 1 convolution) if the number of input channels is
different from the number of output channels. Up2/3d: transposed convolution layers with kernel size (3×)2 × 2 and stride (1×)2 × 2. Proj2d/3d: projection layers (1 × 1 or 1 × 1 × 1
convolutions). The numbers always correspond to the number of channels. With 19 × 19 pixel lenslets (nnum = 19) the rearranged light field input image has 192 = 361 channels. The affine
transformation layer at the end is only part of the network when training on dynamic, single plane targets; otherwise, in inference mode it might be used in post-processing to yield a SPIM
aligned prediction, or the inverse affine transformation is applied to the SPIM target for static samples to avoid unnecessary computations. EXTENDED DATA FIG. 2 LFM-SPIM OPTICAL SETUP.
Schematic 2D drawing of the LFM-SPIM setup showing the main opto-mechanical components. The sample is illuminated through a single illumination objective with two excitation beam paths
(ocra, light sheet illumination and blue, light field selective volume illumination) combined by a dichroic mirror (D1). The fluorescence is detected by an orthogonally oriented detection
objective and optically separated onto two detection arms with a dichroic mirror (D2). Bandpass filters (BP1 and BP2) are placed in front of a tube lens (TL3,TL4) for the respective
detection path. For the light field detection path (green), the tube lens (TL4) focuses on the microlens array (ML) and the image plane (shown in magenta) displaced by one microlens focal
length is relayed by a 1-1 relay lens system (RL6) to an image plane coinciding with the camera sensor (shown in magenta). For the light sheet detection path, a combination of several relay
lenses (RL1 to RL4), a 1:1 macro lens (RL5) together with a lens pair consisting of an offset lens (OL) and an electrically tunable lens (ETL) is used to image two axially displaced
objective focal planes (shown in magenta, dotted and solid) to a common image plane at the sensor. The refocusing is achieved by applying different currents on the ETL. The mirror M1 is
placed at a Fourier plane, such that the FOV of the light sheet path can be laterally aligned to fit the light field detection FOV. For single color imaging, the dichroic mirrors D1 and D2
are replaced by beamsplitters. See Methods for details. EXTENDED DATA FIG. 3 HYLFM-NET PROVIDES ARTIFACT-FREE DECONVOLUTION OF LFM DATA. A, Ground truth single light sheet image. B,
Subdiffraction beads reconstructed by LFM-Net and (C) iterative light field deconvolution (LFD). Scale bar is 10 µm. EXTENDED DATA FIG. 4 PRECISION AND RECALL FOR SUB-DIFFRACTION BEADS.
Precision recall measurements (A-D) and curve (E) for HyLFM-Net-beads, LFD and LFD + CARE. In (A-D) each point represents the average for an individual axial plane. In (E) precision and
recall were averaged over all volumes, such that each point represents a threshold. All reconstructions were scaled to the SPIM ground truth to minimize L2 distance; beads were found
independently using the Difference of Gaussian (DoG) method with varying thresholds and associated with beads found in SPIM (with threshold 0.1) by Hungarian matching. N = 6716 samples were
used in each panel. Note that panel (E) is identical to Fig. 2i. Source data EXTENDED DATA FIG. 5 CROSS-APPLICATION OF TRAINED DEEP NEURAL NETWORKS CAN REVEAL BIAS TO TRAINING DATA. We
created two kinds of samples, one with small (0.1 μm) and one with medium-sized (4 μm) beads suspended in agarose. In (A), HyLFM-Net was trained on small beads and applied to small beads.
FWHM of the beads in the reconstructed volume is shown (6025 beads measured). B, HyLFM-Net was trained on large beads and applied to large beads (682 beads measured). In (C), HyLFM-Net was
trained on small beads and used to reconstruct a volume with large beads (525 beads measured). Similarly, in (D), HyLFM-Net trained on large beads and used to reconstruct a volume with small
beads (2185 beads measured). E, SPIM image of 0.1 μm beads, (F) reconstructions of HyLFM-Net from (A), trained on small beads, (G) reconstructions from HyLFM-Net from (D), trained on large
beads. H, SPIM image of 4 μm beads, I, reconstructions of HyLFM-Net from (B), trained on large beads, (J) reconstructions of HyLFM-Net from (C), trained on small beads. Line profile is shown
to highlight a reconstruction error (red arrows), where the network reconstructs very small beads (as found in the training data) and produces an additional erroneous peak where none is
present in the ground truth SPIM volume. Shadows in (A–D) denote standard deviation. Scale bar 2 μm in (E–G), and 10 μm in (H–J). Source data EXTENDED DATA FIG. 6 NETWORK FINE-TUNING FOR
DIFFERENT DOMAIN GAPS. Refinement of three differently pre-trained HyLFM networks on dynamically acquired medaka heart images. Column (A) shows SPIM ground truth plane at axial position z =
-19µm. Columns (B-D) depict corresponding slices after increasingly many refinement iterations of different pre-trained HyLFM networks. B, A statically trained HyLFM-Net (such as the one in
Fig. 3b), C, HyLFM-Net trained on LFD reconstructions of light-field images acquired on another microscope setup (Wagner, Norlin et al, Nat. Meth. 16, 497–500, 2019) and (D) HyLFM-Net
trained on medium-sized beads (see Fig. 2). E, Respective MS-SSIM image quality metrics for each network and stage of refinement. Standard deviation shown as shadows inferred from N = 756
individual time-points. Note that depending on the domain gap, the refinement converges at different speeds, but high fidelity results can be obtained for all pre-trained networks. Source
data EXTENDED DATA FIG. 7 CA2+-IMAGING IN ZEBRAFISH LARVAE BRAIN. A-C, Representative image plane acquired with SPIM, HyLFM-Net and LFD, respectively (standard deviation projection over
time). D, Selected Ca2+-traces extracted from regions indicated in (A-C), 36 traces with 150 time points analyzed. E, Comparison of Pearson correlation coefficients (R) of Ca2+-traces
extracted by SPIM, HyLFM-Net and LFD. Note that the difference between LFD and HyLFM-Net performance is not statistically significant (p = 0.053, Dunn-Sidak). Scale bar in (A) is 50 µm.
Results representative of n = 5 individual image planes in the volume. Source data SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Tables 1 and 2. REPORTING SUMMARY
SUPPLEMENTARY VIDEO 1 Volumetric HyLFM reconstruction of a beating medaka heart at 40 Hz. Volumetric reconstructions (LFD, HyLFM-Net-stat, and HyLFM-Net-stat refined) of the medaka heart at
40 Hz image acquisition speed shown in Figs. 3 and 4. The cyan plane corresponds to the sweeping SPIM image plane. The panels from left to right show the overlay of the light sheet plane
with the respective plane from the HyLFM-Net/LFD volume, a projection of the HyLFM-Net/LFD volume rotated by 45° around the _y_ axis, and a maximum projection of prediction/reconstruction
volume along the _z_, _y_ and _x_ axes, respectively. Scale bar 30 µm. 41592_2021_1136_MOESM4_ESM.MP4 Supplementary Video 2 Single-plane HyLFM reconstruction of a beating medaka heart at 56
Hz. Single-plane comparison of SPIM ground truth to the corresponding plane of the prediction volume of HyLFM-Net and reconstruction volume of LFD at indicated axial positions of the medaka
heart at 56 Hz image acquisition speed (Fig. 3f,k,m). Scale bar 30 µm. 41592_2021_1136_MOESM5_ESM.MP4 Supplementary Video 3 Single-plane HyLFM reconstruction of a beating medaka heart at 100
Hz. Single-plane comparison of SPIM ground truth to the corresponding plane of the prediction volume of HyLFM-Net at indicated axial positions of the medaka heart at 100 Hz image
acquisition speed. Scale bar 30 µm. 41592_2021_1136_MOESM6_ESM.MP4 Supplementary Video 4 Demonstration of continuous validation and network refinement. Refinement of three differently
pretrained HyLFM networks on dynamically acquired medaka heart images at 40 Hz image acquisition speed. Video of refinement experiments in Supplementary Fig. 6 (see figure caption for
details). SOURCE DATA SOURCE DATA FIG. 2 Statistical source data. SOURCE DATA FIG. 3 Statistical source data. SOURCE DATA FIG. 4 Statistical source data. SOURCE DATA EXTENDED DATA FIG. 4
Precision recall measurements of subdiffraction beads for HyLFM-Net-beads, LFD and LFD + CARE. Source data for Extended Data Fig. 4. SOURCE DATA EXTENDED DATA FIG. 5 Cross-application of
trained deep neural networks and the resulting FWHM of the beads in the reconstructed volume. Source data for Extended Data Fig. 5a–d. SOURCE DATA EXTENDED DATA FIG. 6 MS-SSIM image quality
metrics for each network and stage of refinement. Source data for Extended Data Fig. 6e. SOURCE DATA EXTENDED DATA FIG. 7 Selected Ca2+ traces extracted from HyLFM, LFD and SPIM and
comparison of Pearson correlation coefficients. Source data for Extended Data Fig. 7b,c. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Wagner, N.,
Beuttenmueller, F., Norlin, N. _et al._ Deep learning-enhanced light-field imaging with continuous validation. _Nat Methods_ 18, 557–563 (2021). https://doi.org/10.1038/s41592-021-01136-0
Download citation * Received: 30 July 2020 * Accepted: 01 April 2021 * Published: 07 May 2021 * Issue Date: May 2021 * DOI: https://doi.org/10.1038/s41592-021-01136-0 SHARE THIS ARTICLE
Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided
by the Springer Nature SharedIt content-sharing initiative