Benchmarking single-cell rna-sequencing protocols for cell atlas projects
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
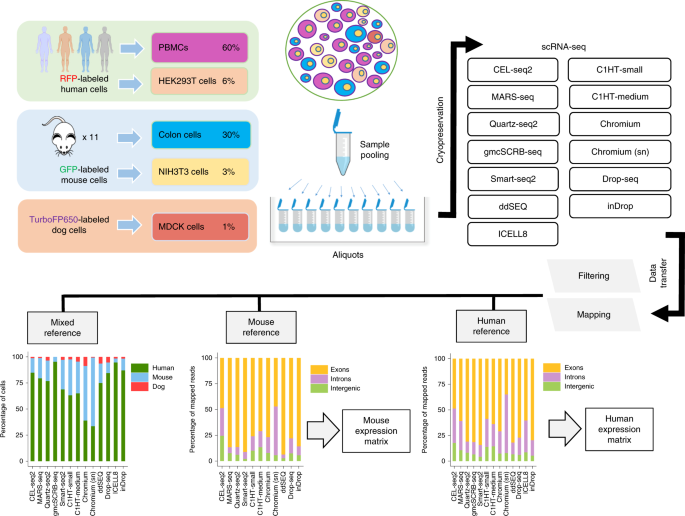
ABSTRACT Single-cell RNA sequencing (scRNA-seq) is the leading technique for characterizing the transcriptomes of individual cells in a sample. The latest protocols are scalable to thousands
of cells and are being used to compile cell atlases of tissues, organs and organisms. However, the protocols differ substantially with respect to their RNA capture efficiency, bias, scale
and costs, and their relative advantages for different applications are unclear. In the present study, we generated benchmark datasets to systematically evaluate protocols in terms of their
power to comprehensively describe cell types and states. We performed a multicenter study comparing 13 commonly used scRNA-seq and single-nucleus RNA-seq protocols applied to a heterogeneous
reference sample resource. Comparative analysis revealed marked differences in protocol performance. The protocols differed in library complexity and their ability to detect cell-type
markers, impacting their predictive value and suitability for integration into reference cell atlases. These results provide guidance both for individual researchers and for consortium
projects such as the Human Cell Atlas. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through
your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this
journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now
Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS RECOVERY OF MISSING SINGLE-CELL RNA-SEQUENCING DATA WITH OPTIMIZED TRANSCRIPTOMIC REFERENCES Article 11 September 2023 A MULTI-CENTER
CROSS-PLATFORM SINGLE-CELL RNA SEQUENCING REFERENCE DATASET Article Open access 02 February 2021 SCANORAMA: INTEGRATING LARGE AND DIVERSE SINGLE-CELL TRANSCRIPTOMIC DATASETS Article 06 June
2024 DATA AVAILABILITY All raw sequencing data and processed gene expression files are freely available through the Gene Expression Omnibus (accession no. GSE133549). CODE AVAILABILITY All
code for the analysis is provided as supplementary material. All code is also available under https://github.com/ati-lz/HCA_Benchmarking and https://github.com/elimereu/matchSCore2.
REFERENCES * Lafzi, A., Moutinho, C., Picelli, S. & Heyn, H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. _Nat. Protoc._ 13, 2742–2757 (2018).
Article CAS PubMed Google Scholar * Prakadan, S. M., Shalek, A. K. & Weitz, D. A. Scaling by shrinking: empowering single-cell ‘omics’ with microfluidic devices. _Nat. Rev. Genet._
18, 345–361 (2017). Article CAS PubMed PubMed Central Google Scholar * Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past
decade. _Nat. Protoc._ 13, 599–604 (2018). Article CAS PubMed Google Scholar * Montoro, D. T. et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. _Nature_
560, 319–324 (2018). Article CAS PubMed PubMed Central Google Scholar * Plasschaert, L. W. et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte.
_Nature_ 560, 377–381 (2018). Article CAS PubMed PubMed Central Google Scholar * Aizarani, N. et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. _Nature_
572, 199–204 (2019). Article CAS PubMed PubMed Central Google Scholar * Karaiskos, N. et al. The _Drosophila_ embryo at single-cell transcriptome resolution. _Science_ 358, 194–199
(2017). Article CAS PubMed Google Scholar * Wagner, D. E. et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. _Science_ 360, 981–987 (2018).
Article CAS PubMed PubMed Central Google Scholar * Regev, A. et al. Science forum: the human cell atlas. _eLife_ 6, e27041 (2017). Article PubMed PubMed Central Google Scholar *
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. _Science_ 357, 661–667 (2017). Article CAS PubMed PubMed Central Google Scholar * Plass,
M. et al. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. _Science_ 360, eaaq1723 (2018). * Moffitt, J. R. et al. High-throughput single-cell
gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. _Proc. Natl Acad. Sci. USA_ 113, 11046–11051 (2016). Article CAS PubMed PubMed Central Google
Scholar * Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M. & Cai, L. Single-cell in situ RNA profiling by sequential hybridization. _Nat. Methods_ 11, 360–361 (2014). Article CAS
PubMed PubMed Central Google Scholar * Alioto, T. S. et al. A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. _Nat. Commun._ 6, 10001
(2015). Article CAS PubMed Google Scholar * Ziegenhain, C. et al. Comparative analysis of single-cell RNA sequencing methods. _Mol. Cell_ 65, 631–643.e4 (2017). Article CAS PubMed
Google Scholar * Svensson, V. et al. Power analysis of single-cell RNA-sequencing experiments. _Nat. Methods_ 14, 381–387 (2017). Article CAS PubMed PubMed Central Google Scholar *
Tung, P.-Y. et al. Batch effects and the effective design of single-cell gene expression studies. _Sci. Rep._ 7, 39921 (2017). Article CAS PubMed PubMed Central Google Scholar * Zheng,
G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. _Nat. Commun._ 8, 14049 (2017). Article CAS PubMed PubMed Central Google Scholar * Haber, A. L. et
al. A single-cell survey of the small intestinal epithelium. _Nature_ 551, 333–339 (2017). Article CAS PubMed PubMed Central Google Scholar * Grün, D. et al. Single-cell messenger RNA
sequencing reveals rare intestinal cell types. _Nature_ 525, 251–255 (2015). Article PubMed CAS Google Scholar * Guillaumet-Adkins, A. et al. Single-cell transcriptome conservation in
cryopreserved cells and tissues. _Genome Biol._ 18, 45 (2017). Article PubMed PubMed Central CAS Google Scholar * Schaum, N. et al. Single-cell transcriptomics of 20 mouse organs
creates a _Tabula Muris_. _Nature_ 562, 367–372 (2018). Article PubMed Central CAS Google Scholar * Büttner, M., Miao, Z., Wolf, F. A., Teichmann, S. A. & Theis, F. J. A test metric
for assessing single-cell RNA-seq batch correction. _Nat. Methods_ 16, 43–49 (2019). Article PubMed CAS Google Scholar * Azuaje, F. A cluster validity framework for genome expression
data. _Bioinforma_ 18, 319–320 (2002). Article CAS Google Scholar * Lin, Y. et al. scMerge leverages factor analysis, stable expression, and pseudoreplication to merge multiple
single-cell RNA-seq datasets. _Proc. Natl Acad. Sci_. _USA_ 116, 9775–9784 (2019). * Kang, H. M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. _Nat.
Biotechnol._ 36, 89–94 (2018). Article CAS PubMed Google Scholar * Stoeckius, M. et al. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell
genomics. _Genome Biol._ 19, 224 (2018). Article CAS PubMed PubMed Central Google Scholar * McGinnis, C. S. et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using
lipid-tagged indices. _Nat. Methods_ 16, 619–626 (2019). Article CAS PubMed PubMed Central Google Scholar * Gaublomme, J. T. et al. Nuclei multiplexing with barcoded antibodies for
single-nucleus genomics. _Nat. Commun._ 10, 1–8 (2019). Article CAS Google Scholar * Mora-Castilla, S. et al. Miniaturization technologies for efficient single-cell library preparation
for next-generation sequencing. _J. Lab. Autom._ 21, 557–567 (2016). Article CAS PubMed PubMed Central Google Scholar * Picelli, S. et al. Tn5 transposase and tagmentation procedures
for massively scaled sequencing projects. _Genome Res._ 24, 2033–2040 (2014). Article CAS PubMed PubMed Central Google Scholar * Brink, S. Cvanden et al. Single-cell sequencing reveals
dissociation-induced gene expression in tissue subpopulations. _Nat. Methods_ 14, 935–936 (2017). Article CAS PubMed Google Scholar * Wohnhaas, C. T. et al. DMSO cryopreservation is the
method of choice to preserve cells for droplet-based single-cell RNA sequencing. _Sci. Rep._ 9, 1–14 (2019). Article CAS Google Scholar * Tosti, L. et al. Single nucleus RNA sequencing
maps acinar cell states in a human pancreas cell atlas. Preprint at _bioRxiv_ https://doi.org/10.1101/733964 (2019). * Massoni-Badosa, R. et al. Sampling artifacts in single-cell genomics
cohort studies. Preprint at _bioRxiv_ https://doi.org/10.1101/2020.01.15.897066 (2020). * Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells.
_Nat. Methods_ 10, 1096–1098 (2013). Article CAS PubMed Google Scholar * Sasagawa, Y. et al. Quartz-Seq2: a high-throughput single-cell RNA-sequencing method that effectively uses
limited sequence reads. _Genome Biol._ 19, 29 (2018). Article PubMed PubMed Central CAS Google Scholar * Hashimshony, T. et al. CEL-Seq2: sensitive highly-multiplexed single-cell
RNA-Seq. _Genome Biol._ 17, 77 (2016). Article PubMed PubMed Central CAS Google Scholar * Bagnoli, J. W. et al. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. _Nat.
Commun._ 9, 2937 (2018). Article PubMed PubMed Central CAS Google Scholar * Sasagawa, Y. et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method,
reveals non-genetic gene-expression heterogeneity. _Genome Biol._ 14, 3097 (2013). Article CAS Google Scholar * Parekh, S., Ziegenhain, C., Vieth, B., Enard, W. & Hellmann, I. The
impact of amplification on differential expression analyses by RNA-seq. _Sci. Rep._ 6, 25533 (2016). Article CAS PubMed PubMed Central Google Scholar * Soneson, C. & Robinson, M. D.
Bias, robustness and scalability in single-cell differential expression analysis. _Nat. Methods_ 15, 255–261 (2018). Article CAS PubMed Google Scholar * Saelens, W. et al. A comparison
of single-cell trajectory inference methods. _Nat. Biotechnol._ 37, 547–554 (2019). Article CAS PubMed Google Scholar * Holland, C. H. et al. Robustness and applicability of
transcription factor and pathway analysis tools on single-cell RNA-seq data. _Genome Biol._ 21, 36 (2020). Article CAS PubMed PubMed Central Google Scholar * Kharchenko, P. V.,
Silberstein, L. & Scadden, D. T. Bayesian approach to single-cell differential expression analysis. _Nat. Methods_ 11, 740–742 (2014). Article CAS PubMed PubMed Central Google
Scholar * Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. _Nat. Biotechnol._ 33, 495–502 (2015). Article
CAS PubMed PubMed Central Google Scholar * Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. _Nat. Methods_ 16, 1289–1296 (2019). Article
CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This project has been made possible in part by grant no. 2018-182827 from the Chan Zuckerberg Initiative
DAF, an advised fund of the Silicon Valley Community Foundation. H.H. is a Miguel Servet (CP14/00229) researcher funded by the Spanish Institute of Health Carlos III (ISCIII). C.M. is
supported by an AECC postdoctoral fellowship. This work has received funding from the European Union’s Horizon 2020 research and innovation program under Marie Skłodowska-Curie grant
agreement no. H2020-MSCA-ITN-2015-675752 (Singek), and the Ministerio de Ciencia, Innovación y Universidades (SAF2017-89109-P; AEI/FEDER, UE). S. was supported by the German Research
Foundation’s (DFG’s) (GR4980) Behrens-Weise-Foundation. D.G. and S. are supported by the Max Planck Society. C.Z. was supported by the European Molecular Biology Organization through the
long-term fellowship ALTF 673-2017. The snRNA-seq data were generated with support from the National Institute of Allergy and Infectious Diseases (grant no. U24AI118672), the Manton
Foundation and the Klarman Cell Observatory (to A.R.). I.N. was supported by JST CREST (grant no. JPMJCR16G3), Japan, and the Projects for Technological Development, Research Center Network
for Realization of Regenerative Medicine by Japan, the Japan Agency for Medical Research and Development. A.J., L.E.W., J.W.B. and W.E. were supported by funding from the DFG (EN 1093/2-1
and SFB1243 TP A14). We thank ThePaperMill for critical reading and scientific editing services and the Eukaryotic Single Cell Genomics Facility at Scilifelab (Stockholm, Sweden) for
support. This publication is part of a project (BCLLATLAS) that received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program
(grant agreement no. 810287). Core funding was from the ISCIII and the Generalitat de Catalunya. AUTHOR INFORMATION Author notes * These authors contributed equally: Elisabetta Mereu, Atefeh
Lafzi. AUTHORS AND AFFILIATIONS * CNAG-CRG, Centre for Genomic Regulation, Barcelona Institute of Science and Technology, Barcelona, Spain Elisabetta Mereu, Atefeh Lafzi, Catia Moutinho,
Marta Gut, Ivo Gut & Holger Heyn * Department of Cell and Molecular Biology, Karolinska Institutet, Stockholm, Sweden Christoph Ziegenhain & Rickard Sandberg * European Molecular
Biology Laboratory, European Bioinformatics Institute, Hinxton, Cambridge, UK Davis J. McCarthy & Oliver Stegle * European Molecular Biology Laboratory, Genome Biology Unit, Heidelberg,
Germany Davis J. McCarthy & Oliver Stegle * St Vincent’s Institute of Medical Research, Fitzroy, Victoria, Australia Davis J. McCarthy * Institute for Research in Biomedicine, Barcelona
Institute of Science and Technology, Barcelona, Spain Adrián Álvarez-Varela & Eduard Batlle * Catalan Institution for Research and Advanced Studies, Barcelona, Spain Eduard Batlle *
Centro de Investigación Biomédica en Red de Cáncer, Barcelona, Spain Eduard Batlle * Max-Planck-Institute of Immunobiology and Epigenetics, Freiburg, Germany Sagar & Dominic Grün * 10x
Genomics, Pleasanton, CA, USA Julia K. Lau & Stéphane C. Boutet * Fluidigm Corporation, South San Francisco, CA, USA Chad Sanada & Aik Ooi * Department of Bioengineering, Stanford
University, Stanford, CA, USA Robert C. Jones * Bio-Rad, Hercules, CA, USA Kelly Kaihara, Chris Brampton & Yasha Talaga * Laboratory for Bioinformatics Research, RIKEN Center for
Biosystems, Dynamics Research, Saitama, Japan Yohei Sasagawa, Kaori Tanaka, Tetsutaro Hayashi & Itoshi Nikaido * Max Delbrück Center for Molecular Medicine/Berlin Institute of Health,
Berlin, Germany Caroline Braeuning, Cornelius Fischer & Sascha Sauer * Digital Health Center, Berlin Institute of Health, Charité-Universitätsmedizin Berlin, Berlin, Germany Timo Trefzer
& Christian Conrad * Klarman Cell Observatory, Broad Institute of MIT and Harvard, Cambridge, MA, USA Xian Adiconis, Lan T. Nguyen, Aviv Regev & Joshua Z. Levin * Stanley Center for
Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA Xian Adiconis & Joshua Z. Levin * Koch Institute of Integrative Cancer Research, MIT, Cambridge, MA, USA
Aviv Regev * Howard Hughes Medical Institute, Department of Biology, MIT, Cambridge, MA, USA Aviv Regev * Max-Planck-Institute for Biology of Ageing, Cologne, Germany Swati Parekh *
Anthropology & Human Genomics, Department of Biology II, Ludwig-Maximilians-University, Martinsried, Germany Aleksandar Janjic, Lucas E. Wange, Johannes W. Bagnoli & Wolfgang Enard *
School of Integrative and Global Majors, University of Tsukuba, Wako, Saitama, Japan Itoshi Nikaido * Universitat Pompeu Fabra, Barcelona, Spain Ivo Gut & Holger Heyn * Division of
Computational Genomics and Systems Genetics, German Cancer Research Center, Heidelberg, Germany Oliver Stegle Authors * Elisabetta Mereu View author publications You can also search for this
author inPubMed Google Scholar * Atefeh Lafzi View author publications You can also search for this author inPubMed Google Scholar * Catia Moutinho View author publications You can also
search for this author inPubMed Google Scholar * Christoph Ziegenhain View author publications You can also search for this author inPubMed Google Scholar * Davis J. McCarthy View author
publications You can also search for this author inPubMed Google Scholar * Adrián Álvarez-Varela View author publications You can also search for this author inPubMed Google Scholar * Eduard
Batlle View author publications You can also search for this author inPubMed Google Scholar * Sagar View author publications You can also search for this author inPubMed Google Scholar *
Dominic Grün View author publications You can also search for this author inPubMed Google Scholar * Julia K. Lau View author publications You can also search for this author inPubMed Google
Scholar * Stéphane C. Boutet View author publications You can also search for this author inPubMed Google Scholar * Chad Sanada View author publications You can also search for this author
inPubMed Google Scholar * Aik Ooi View author publications You can also search for this author inPubMed Google Scholar * Robert C. Jones View author publications You can also search for this
author inPubMed Google Scholar * Kelly Kaihara View author publications You can also search for this author inPubMed Google Scholar * Chris Brampton View author publications You can also
search for this author inPubMed Google Scholar * Yasha Talaga View author publications You can also search for this author inPubMed Google Scholar * Yohei Sasagawa View author publications
You can also search for this author inPubMed Google Scholar * Kaori Tanaka View author publications You can also search for this author inPubMed Google Scholar * Tetsutaro Hayashi View
author publications You can also search for this author inPubMed Google Scholar * Caroline Braeuning View author publications You can also search for this author inPubMed Google Scholar *
Cornelius Fischer View author publications You can also search for this author inPubMed Google Scholar * Sascha Sauer View author publications You can also search for this author inPubMed
Google Scholar * Timo Trefzer View author publications You can also search for this author inPubMed Google Scholar * Christian Conrad View author publications You can also search for this
author inPubMed Google Scholar * Xian Adiconis View author publications You can also search for this author inPubMed Google Scholar * Lan T. Nguyen View author publications You can also
search for this author inPubMed Google Scholar * Aviv Regev View author publications You can also search for this author inPubMed Google Scholar * Joshua Z. Levin View author publications
You can also search for this author inPubMed Google Scholar * Swati Parekh View author publications You can also search for this author inPubMed Google Scholar * Aleksandar Janjic View
author publications You can also search for this author inPubMed Google Scholar * Lucas E. Wange View author publications You can also search for this author inPubMed Google Scholar *
Johannes W. Bagnoli View author publications You can also search for this author inPubMed Google Scholar * Wolfgang Enard View author publications You can also search for this author
inPubMed Google Scholar * Marta Gut View author publications You can also search for this author inPubMed Google Scholar * Rickard Sandberg View author publications You can also search for
this author inPubMed Google Scholar * Itoshi Nikaido View author publications You can also search for this author inPubMed Google Scholar * Ivo Gut View author publications You can also
search for this author inPubMed Google Scholar * Oliver Stegle View author publications You can also search for this author inPubMed Google Scholar * Holger Heyn View author publications You
can also search for this author inPubMed Google Scholar CONTRIBUTIONS H.H. designed the study. E.M. and A.L. performed all data analyses. C.M., A.A.V. and E.B. prepared the reference
sample. C.Z., D.J.M., S.P. and O.S. supported the data analysis. M.G. and I.G. provided technical and sequencing support. S., D.G., J.K.L., S.C.B., C.S., A.O., R.C.J., K.K., C.B., Y.T.,
Y.S., K.T., T.H., C.B., C.F., S.S., T.T., C.C., X.A., L.T.N., A.R., J.Z.L., A.J., L.E.W., J.W.B., W.E., R.S. and I.N. provided sequencing-ready single-cell libraries or sequencing raw data.
H.H., E.M. and A.L. wrote the manuscript with contributions from the co-authors. All authors read and approved the final manuscript. CORRESPONDING AUTHOR Correspondence to Holger Heyn.
ETHICS DECLARATIONS COMPETING INTERESTS A.R. is a co-founder and equity holder of Celsius Therapeutics, and an SAB member of Thermo Fisher Scientific and Syros Pharmaceuticals. He is also a
co-inventor on patent applications to numerous advances in single-cell genomics, including droplet-based sequencing technologies, as in PCT/US2015/0949178, and methods for expression and
analysis, as in PCT/US2016/059233 and PCT/US2016/059239. K.K., C.B. and Y.T. are employed by Bio-Rad Laboratories. J.K.L. and S.C.B. are employees and shareholders at 10x Genomics, Inc.
S.C.B. is a former employee and shareholder of Fluidigm Corporation. C.S. and A.O. are employed by Fluidigm. All other authors declare no conflicts of interest associated with this
manuscript. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION Supplementary Figs. 1–31 and Supplementary Notes. REPORTING SUMMARY. SUPPLEMENTARY TABLE 1 SUPPLEMENTARY TABLE 2 SUPPLEMENTARY TABLE 3 SUPPLEMENTARY
TABLE 4 SUPPLEMENTARY TABLE 5 SUPPLEMENTARY TABLE 6 SUPPLEMENTARY TABLE 7 SUPPLEMENTARY TABLE 8 SUPPLEMENTARY CODE RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE
THIS ARTICLE Mereu, E., Lafzi, A., Moutinho, C. _et al._ Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. _Nat Biotechnol_ 38, 747–755 (2020).
https://doi.org/10.1038/s41587-020-0469-4 Download citation * Received: 07 May 2019 * Revised: 18 February 2020 * Accepted: 26 February 2020 * Published: 06 April 2020 * Issue Date: June
2020 * DOI: https://doi.org/10.1038/s41587-020-0469-4 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative