Mhc-ii neoantigens shape tumour immunity and response to immunotherapy
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The ability of the immune system to eliminate and shape the immunogenicity of tumours defines the process of cancer immunoediting1. Immunotherapies such as those that target immune
checkpoint molecules can be used to augment immune-mediated elimination of tumours and have resulted in durable responses in patients with cancer that did not respond to previous treatments.
However, only a subset of patients benefit from immunotherapy and more knowledge about what is required for successful treatment is needed2,3,4. Although the role of tumour
neoantigen-specific CD8+ T cells in tumour rejection is well established5,6,7,8,9, the roles of other subsets of T cells have received less attention. Here we show that spontaneous and
immunotherapy-induced anti-tumour responses require the activity of both tumour-antigen-specific CD8+ and CD4+ T cells, even in tumours that do not express major histocompatibility complex
(MHC) class II molecules. In addition, the expression of MHC class II-restricted antigens by tumour cells is required at the site of successful rejection, indicating that activation of CD4+
T cells must also occur in the tumour microenvironment. These findings suggest that MHC class II-restricted neoantigens have a key function in the anti-tumour response that is nonoverlapping
with that of MHC class I-restricted neoantigens and therefore needs to be considered when identifying patients who will most benefit from immunotherapy. Access through your institution Buy
or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get
Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 51 print issues and online access $199.00 per year only $3.90
per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout
ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS ANTIGEN PRESENTATION IN
CANCER: INSIGHTS INTO TUMOUR IMMUNOGENICITY AND IMMUNE EVASION Article 09 March 2021 CD4+ T CELL-INDUCED INFLAMMATORY CELL DEATH CONTROLS IMMUNE-EVASIVE TUMOURS Article Open access 14 June
2023 PHENOTYPE, SPECIFICITY AND AVIDITY OF ANTITUMOUR CD8+ T CELLS IN MELANOMA Article 21 July 2021 DATA AVAILABILITY Nucleotide variant calls generated from cDNA capture sequencing of the
T3 and KP9025 sarcoma lines and used in the prediction of antigens shown in Fig. 1a, Extended Data Fig. 3a, b and 6b are provided as Supplementary Data 1 and Supplementary Data 2. CODE
AVAILABILITY Code for the hmMHC algorithm used to predict presentation of neoantigens by I-Ab can be accessed at https://github.com/artyomovlab/hmmhc. REFERENCES * Schreiber, R. D., Old, L.
J. & Smyth, M. J. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. _Science_ 331, 1565–1570 (2011). Article ADS CAS PubMed Google Scholar *
Larkin, J. et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. _N. Engl. J. Med_. 373, 23–34 (2015). Article PubMed PubMed Central CAS Google Scholar *
Motzer, R. J. et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. _N. Engl. J. Med_. 373, 1803–1813 (2015). Article CAS PubMed PubMed Central Google Scholar * Borghaei,
H. et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. _N. Engl. J. Med_. 373, 1627–1639 (2015). Article CAS PubMed PubMed Central Google Scholar *
Lennerz, V. et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. _Proc. Natl Acad. Sci. USA_ 102, 16013–16018 (2005). Article ADS CAS PubMed
PubMed Central Google Scholar * Matsushita, H. et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. _Nature_ 482, 400–404 (2012). Article ADS CAS
PubMed PubMed Central Google Scholar * DuPage, M., Mazumdar, C., Schmidt, L. M., Cheung, A. F. & Jacks, T. Expression of tumour-specific antigens underlies cancer immunoediting.
_Nature_ 482, 405–409 (2012). Article ADS CAS PubMed PubMed Central Google Scholar * Robbins, P. F. et al. Mining exomic sequencing data to identify mutated antigens recognized by
adoptively transferred tumor-reactive T cells. _Nat. Med_. 19, 747–752 (2013). Article CAS PubMed PubMed Central Google Scholar * Gubin, M. M. et al. Checkpoint blockade cancer
immunotherapy targets tumour-specific mutant antigens. _Nature_ 515, 577–581 (2014). Article ADS CAS PubMed PubMed Central Google Scholar * Wölfel, T. et al. A p16INK4a-insensitive
CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. _Science_ 269, 1281–1284 (1995). Article ADS PubMed Google Scholar * Snyder, A. et al. Genetic basis for clinical
response to CTLA-4 blockade in melanoma. _N. Engl. J. Med_. 371, 2189–2199 (2014). Article PubMed PubMed Central CAS Google Scholar * Strønen, E. et al. Targeting of cancer neoantigens
with donor-derived T cell receptor repertoires. _Science_ 352, 1337–1341 (2016). Article ADS PubMed CAS Google Scholar * Rizvi, N. A. et al. Mutational landscape determines sensitivity
to PD-1 blockade in non-small cell lung cancer. _Science_ 348, 124–128 (2015). Article ADS CAS PubMed PubMed Central Google Scholar * Van Allen, E. M. et al. Genomic correlates of
response to CTLA-4 blockade in metastatic melanoma. _Science_ 350, 207–211 (2015). Article ADS PubMed PubMed Central CAS Google Scholar * Spranger, S. et al. Density of immunogenic
antigens does not explain the presence or absence of the T-cell-inflamed tumor microenvironment in melanoma. _Proc. Natl Acad. Sci. USA_ 113, E7759–E7768 (2016). Article CAS PubMed PubMed
Central Google Scholar * Hugo, W. et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. _Cell_ 165, 35–44 (2016). Article CAS PubMed
PubMed Central Google Scholar * Hellmann, M. D. et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. _Cancer Cell_ 33,
843–852 (2018). Article CAS PubMed PubMed Central Google Scholar * Kreiter, S. et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. _Nature_ 520, 692–696
(2015). Article ADS CAS PubMed PubMed Central Google Scholar * Ott, P. A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. _Nature_ 547, 217–221 (2017).
Article ADS CAS PubMed PubMed Central Google Scholar * Ossendorp, F., Mengedé, E., Camps, M., Filius, R. & Melief, C. J. Specific T helper cell requirement for optimal induction of
cytotoxic T lymphocytes against major histocompatibility complex class II negative tumors. _J. Exp. Med_. 187, 693–702 (1998). Article CAS PubMed PubMed Central Google Scholar *
Corthay, A. et al. Primary antitumor immune response mediated by CD4+ T cells. _Immunity_ 22, 371–383 (2005). Article CAS PubMed Google Scholar * Wong, S. B. J., Bos, R. & Sherman,
L. A. Tumor-specific CD4+ T cells render the tumor environment permissive for infiltration by low-avidity CD8+ T cells. _J. Immunol_. 180, 3122–3131 (2008). Article CAS PubMed Google
Scholar * Bos, R. & Sherman, L. A. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. _Cancer Res_. 70, 8368–8377 (2010).
Article CAS PubMed PubMed Central Google Scholar * Zhu, Z. et al. CD4+ T cell help selectively enhances high-avidity tumor antigen-specific CD8+ T cells. _J. Immunol_. 195, 3482–3489
(2015). Article CAS PubMed Google Scholar * Borst, J., Ahrends, T., Bąbała, N., Melief, C. J. M. & Kastenmüller, W. CD4+ T cell help in cancer immunology and immunotherapy. _Nat.
Rev. Immunol_. 18, 635–647 (2018). Article CAS PubMed Google Scholar * Andreatta, M. et al. An automated benchmarking platform for MHC class II binding prediction methods.
_Bioinformatics_ 34, 1522–1528 (2018). Article CAS PubMed Google Scholar * Mittal, P. et al. Tumor-unrelated CD4 T cell help augments CD134 plus CD137 dual costimulation tumor therapy.
_J. Immunol_. 195, 5816–5826 (2015). Article CAS PubMed Google Scholar * Suri, A., Walters, J. J., Rohrs, H. W., Gross, M. L. & Unanue, E. R. First signature of islet β-cell-derived
naturally processed peptides selected by diabetogenic class II MHC molecules. _J. Immunol_. 180, 3849–3856 (2008). Article CAS PubMed Google Scholar * Tran, E. et al. Cancer
immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. _Science_ 344, 641–645 (2014). Article ADS CAS PubMed PubMed Central Google Scholar *
Linnemann, C. et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. _Nat. Med_. 21, 81–85 (2015). Article CAS PubMed
Google Scholar * Old, L. J. & Boyse, E. A. Immunology of experimental tumors. _Annu. Rev. Med_. 15, 167–186 (1964). Article CAS PubMed Google Scholar * Wei, S. C. et al. Distinct
cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. _Cell_ 170, 1120–1133 (2017). Article CAS PubMed PubMed Central Google Scholar * Gubin, M. M. et al.
High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. _Cell_ 175, 1014–1030 (2018). Article CAS PubMed
PubMed Central Google Scholar * Marzo, A. L. et al. Tumor-specific CD4+ T cells have a major “post-licensing” role in CTL mediated anti-tumor immunity. _J. Immunol_. 165, 6047–6055 (2000).
Article CAS PubMed Google Scholar * Bennett, S. R. M., Carbone, F. R., Karamalis, F., Miller, J. F. A. P. & Heath, W. R. Induction of a CD8+ cytotoxic T lymphocyte response by
cross-priming requires cognate CD4+ T cell help. _J. Exp. Med_. 186, 65–70 (1997). Article CAS PubMed PubMed Central Google Scholar * Corthay, A., Lundin, K. U., Lorvik, K. B.,
Hofgaard, P. O. & Bogen, B. Secretion of tumor-specific antigen by myeloma cells is required for cancer immunosurveillance by CD4+ T cells. _Cancer Res_. 69, 5901–5907 (2009). Article
CAS PubMed Google Scholar * Abelin, J. G. et al. Defining HLA-II ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. _Immunity_
https://doi.org/10.1016/j.immuni.2019.08.012 (2019). Article CAS PubMed Google Scholar * Mamitsuka, H. Predicting peptides that bind to MHC molecules using supervised learning of hidden
Markov models. _Proteins_ 33, 460–474 (1998). Article CAS PubMed Google Scholar * Welch, L. R. Hidden Markov models and the Baum–Welch algorithm. _IEEE Inf. Theory Soc. Newsl_. 53, 10–13
(2003). Google Scholar * Jurtz, V. et al. NetMHCpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. _J. Immunol_.
199, 3360–3368 (2017). Article CAS PubMed Google Scholar * Jensen, K. K. et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. _Immunology_ 154,
394–406 (2018). Article CAS PubMed PubMed Central Google Scholar * Kowalewski, D. J. & Stevanović, S. Biochemical large-scale identification of MHC class I ligands. _Methods Mol.
Biol_. 960, 145–157 (2013). Article CAS PubMed Google Scholar * Tungatt, K. et al. Antibody stabilization of peptide-MHC multimers reveals functional T cells bearing extremely
low-affinity TCRs. _J. Immunol_. 194, 463–474 (2015). Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank all members of the Schreiber laboratory for
discussions and technical support. This work was supported by grants to R.D.S. from the National Cancer Institute of the National Institutes of Health (RO1CA190700), the Parker Institute for
Cancer Immunotherapy, the Cancer Research Institute, Janssen Pharmaceutical Company of Johnson and Johnson and the Prostate Cancer Foundation, and by a Stand Up to Cancer-Lustgarten
Foundation Pancreatic Cancer Foundation Convergence Dream Team Translational Research Grant. Stand Up to Cancer is a program of the Entertainment Industry Foundation administered by the
American Association for Cancer Research. E.A. and D.M.L were supported by a postdoctoral training grant (T32 CA00954729) from the National Cancer Institute. D.M.L. and M.M.G. were supported
by the Irvington Postdoctoral Fellowship from the Cancer Research Institute. M.D. is a St Baldrick’s Scholar with support from Hope with Hazel and a Pew-Stewart Scholar for Cancer Research
supported by the Pew Charitable Trusts. J.P.W. is supported by the National Cancer Institute of the National Institutes of Health Paul Calabresi Career Development Award in Clinical Oncology
(K12CA167540). M.M.G. is supported by a Parker Bridge Scholar Award from the Parker Institute for Cancer Immunotherapy. K.W.W. receives support from the National Institutes of Health
(R01CA238039). T.J. receives support from a National Institutes of Health Cancer Center Support Grant (P30CA14051) and the Howard Hughes Medical Institute. E.R.U. receives support from the
National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (AI114551 and DK058177). Aspects of the studies, including ELISPOT, were performed by D.
Bender at the Immunomonitoring Laboratory (IML), which is supported by the Andrew M. and Jane N. Bursky Center for Human Immunology and Immunotherapy Programs and the Alvin J. Siteman
Comprehensive Cancer Center which, in turn, is supported by a National Cancer Institute of the National Institutes of Health Cancer Center Support Grant (P30CA91842). AUTHOR INFORMATION
Author notes * Michel DuPage Present address: Division of Immunology and Pathogenesis, Department of Molecular and Cell Biology, University of California Berkeley, Berkeley, CA, USA AUTHORS
AND AFFILIATIONS * Department of Pathology and Immunology, Washington University School of Medicine, St Louis, MO, USA Elise Alspach, Danielle M. Lussier, Alexander P. Miceli, Ilya
Kizhvatov, Wei Meng, Cheryl F. Lichti, Ekaterina Esaulova, Anthony N. Vomund, Daniele Runci, Jeffrey P. Ward, Matthew M. Gubin, Ruan F. V. Medrano, Cora D. Arthur, J. Michael White, Kathleen
C. F. Sheehan, Alex Chen, Emil R. Unanue, Maxim N. Artyomov & Robert D. Schreiber * The Andrew M. and Jane M. Bursky Center for Human Immunology and Immunotherapy Programs, Washington
University School of Medicine, St Louis, MO, USA Elise Alspach, Danielle M. Lussier, Alexander P. Miceli, Wei Meng, Cheryl F. Lichti, Daniele Runci, Jeffrey P. Ward, Matthew M. Gubin, Ruan
F. V. Medrano, Cora D. Arthur, Kathleen C. F. Sheehan & Robert D. Schreiber * David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge,
MA, USA Michel DuPage & Tyler Jacks * Department of Cancer Immunology and Virology, Dana-Farber Cancer Institute, Boston, MA, USA Adrienne M. Luoma & Kai W. Wucherpfennig * Division
of Oncology, Department of Medicine, Washington University School of Medicine, St Louis, MO, USA Jeffrey P. Ward * Howard Hughes Medical Institute, Massachusetts Institute of Technology,
Cambridge, MA, USA Tyler Jacks * The Parker Institute for Cancer Immunotherapy, San Francisco, CA, USA Robert D. Schreiber Authors * Elise Alspach View author publications You can also
search for this author inPubMed Google Scholar * Danielle M. Lussier View author publications You can also search for this author inPubMed Google Scholar * Alexander P. Miceli View author
publications You can also search for this author inPubMed Google Scholar * Ilya Kizhvatov View author publications You can also search for this author inPubMed Google Scholar * Michel DuPage
View author publications You can also search for this author inPubMed Google Scholar * Adrienne M. Luoma View author publications You can also search for this author inPubMed Google Scholar
* Wei Meng View author publications You can also search for this author inPubMed Google Scholar * Cheryl F. Lichti View author publications You can also search for this author inPubMed
Google Scholar * Ekaterina Esaulova View author publications You can also search for this author inPubMed Google Scholar * Anthony N. Vomund View author publications You can also search for
this author inPubMed Google Scholar * Daniele Runci View author publications You can also search for this author inPubMed Google Scholar * Jeffrey P. Ward View author publications You can
also search for this author inPubMed Google Scholar * Matthew M. Gubin View author publications You can also search for this author inPubMed Google Scholar * Ruan F. V. Medrano View author
publications You can also search for this author inPubMed Google Scholar * Cora D. Arthur View author publications You can also search for this author inPubMed Google Scholar * J. Michael
White View author publications You can also search for this author inPubMed Google Scholar * Kathleen C. F. Sheehan View author publications You can also search for this author inPubMed
Google Scholar * Alex Chen View author publications You can also search for this author inPubMed Google Scholar * Kai W. Wucherpfennig View author publications You can also search for this
author inPubMed Google Scholar * Tyler Jacks View author publications You can also search for this author inPubMed Google Scholar * Emil R. Unanue View author publications You can also
search for this author inPubMed Google Scholar * Maxim N. Artyomov View author publications You can also search for this author inPubMed Google Scholar * Robert D. Schreiber View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS E.A. conceived and designed the experiments, collected the data, performed and interpreted the
analyses, and wrote the manuscript. D.M.L. and A.P.M. planned experiments, and collected and analysed data. I.K. conceived of and designed the hmMHC algorithm and performed analyses using
it, and wrote the methodological description. M.D. generated the KP9025 sarcoma cell line. A.M.L provided technical assistance and helped to plan experiments using MHC-II tetramers. W.M. and
C.F.L. planned, performed and analysed mass spectrometry experiments. E.E. assisted with bioinformatics analyses. A.N.V. assisted with the generation of the CD4+ T cell hybridomas, and
helped to design and perform experiments using them. D.R. designed experiments involving multi-colour flow cytometry and collected and analysed the data. J.P.W. provided technical support
for MHC-I tetramer staining. M.M.G. assisted in experiment planning. R.F.V.M. collected and analysed data for experiments involving multi-colour flow cytometry. C.D.A., K.C.F.S. and J.M.W.
provided technical assistance throughout the study. A.C. collected data. K.W.W. provided mITGB1–MHC-II monomers and provided assistance in experimental design. T.J. provided support in
experimental design and data analysis regarding the KP9025 sarcoma line. M.N.A. conceived and designed the hmMHC algorithm and provided bioinformatics support. E.R.U. provided assistance
with experimental design. R.D.S. conceived experiments, interpreted data, and wrote the manuscript. All authors contributed to manuscript revision. CORRESPONDING AUTHOR Correspondence to
Robert D. Schreiber. ETHICS DECLARATIONS COMPETING INTERESTS R.D.S. is a cofounder, scientific advisory board member, stockholder, and royalty recipient of Jounce Therapeutics and Neon
Therapeutics and is a scientific advisory board member for A2 Biotherapeutics, BioLegend, Codiak Biosciences, Constellation Pharmaceuticals, NGM Biopharmaceuticals and Sensei
Biotherapeutics. K.W.W. serves on the scientific advisory board of Tscan Therapeutics and Nextechinvest, and receives sponsored research funding from Bristol-Myers Squibb and Novartis; these
activities are not related to the findings described in this publication. T.J. is a member of the Board of Directors of Amgen and Thermo Fisher Scientific. He is also a co-founder of
Dragonfly Therapeutics and T2 Biosystems, and serves on the Scientific Advisory Board of Dragonfly Therapeutics, SQZ Biotech, and Skyhawk Therapeutics. None of these affiliations represent a
conflict of interest with respect to the design or execution of this study or interpretation of data presented in this manuscript. The laboratory of T.J. currently also receives funding
from the Johnson & Johnson Lung Cancer Initiative and Calico, but this funding did not support the research described in this manuscript. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer
Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. PEER REVIEW INFORMATION _Nature_ thanks Lelia Delamarre, Cornelis Melief and
the other, anonymous, reviewer(s) for their contribution to the peer review of this work. EXTENDED DATA FIGURES AND TABLES EXTENDED DATA FIG. 1 THE HMMHC PREDICTIVE ALGORITHM AND IEDB′18
H2-I-AB TRAINING DATASET COMPOSITION. A, An example of a fully connected HMM with three hidden states, and emissions corresponding to amino acids. B–D, Composition of IEDB dataset (MHC full
ligand export downloaded on 25 November 2018) represented as number of peptides per binding category and measurement type (B, C) and binding category and peptide length (D). Strong binders:
IC50 ≤ 50 nM; binders: 50 nM < IC50 ≤ 500 nM; weak binders: 500 nM < IC50 ≤ 5,000 nM; non-binders: all remaining peptides. Source data EXTENDED DATA FIG. 2 PERFORMANCE OF HMMHC
COMPARED TO NETMHCII-2.3 AND NETMHCIIPAN-3.2. A, hmMHC (orange shapes) underwent 10× cross-validation. In each of the ten cross-validation partitions, on average there were 4,412 binders in
the training set, and 771 binders and 77,086 random natural peptides in the validation set. Performance was compared in terms of AUROC to the performance of netMHCII-2.3 (blue triangles) and
netMHCIIpan-3.2 (purple triangles) applied on the same validation sets. For hmMHC, performance for different numbers of hidden states is shown. For netMHCII-2.3 and netMHCIIpan-3.2,
performance is shown for both predicted affinity and percentile rank (PR). B, Receiver operating characteristic (ROC) curves showing the performance of hmMHC on the H2-I-Ab dataset compared
to existing predictors. ROC curves of all peptides and per specific peptide length for every cross-validation partition are shown. C, Illustration of percentile rank for strong binder
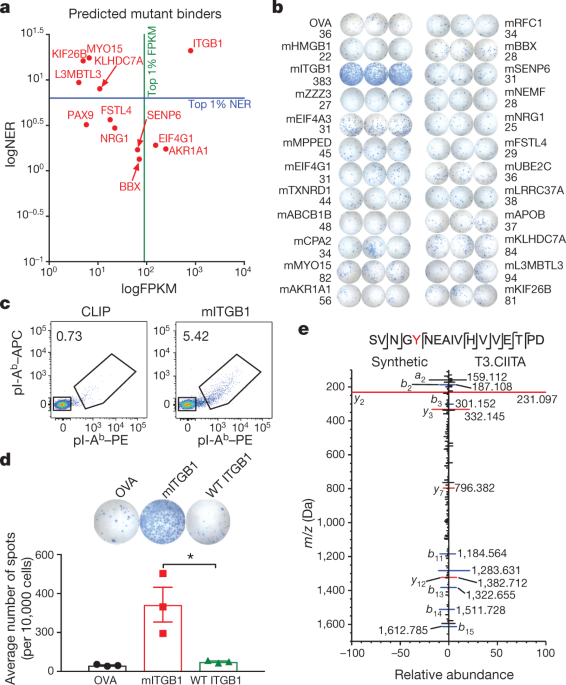
classification calibrated on random natural peptides. Red lines indicate the percentile ranks of peptides screened for CD4+ T cell reactivity. Source data EXTENDED DATA FIG. 3 MITGB1 IS A
MAJOR MHC-II-RESTRICTED NEOANTIGEN IN T3 SARCOMAS. A, B, T3 MHC-II neoantigen predictions for all expressed mutations were made using hmMHC (A) and netMHCII-2.3 (B) (netMHCIIpan-3.2
predictions yielded very similar results, data not shown). The predictions are shown as −log10odds predictor value or logIC50 (smaller values indicate higher likelihood of being presented by
I-Ab) and expression level (FPKM). Strong binders are defined as mutations residing in the second percentile of I-Ab binding predictions for random natural peptides for each algorithm
(−log10odds ≤ 26.21 or logIC50 ≤ 343.8 nM). The N710Y mutation in ITGB1 met the strong binder threshold in the hmMHC predictions but not in the netMHCII-2.3 predictions. Red dots indicate
all mutations that were screened for CD4+ T cell reactivity. Green line denotes high-expression cut-off (FPKM = 89.1). Blue line indicates strong binder cut off for each algorithm. C, Two
million T3 sarcoma cells were injected subcutaneously into syngeneic mice and CD4+ TILs were isolated on day 12. IFNγ ELISPOT was performed using naive splenocytes pulsed with 2 μg ml−1 of
the indicated peptides. Data are shown as mean of three independent experiments ± s.e.m. D, Gating strategy for pI-Ab tetramer staining of whole TILs. E, Quantification of mITGB1–tetramer
and CLIP–tetramer staining of CD4+ T cells from whole T3 TILs 12 days after transplantation. Data are shown as mean ± s.e.m. per cent tetramer-positive cells of CD4+ cells from three
independent experiments. F, Syngeneic 129S6 mice were injected subcutaneously with 2 × 106 T3 sarcoma cells and TIL-derived CD4+ T cells were collected 12 days after transplantation. CD4+ T
cells were stimulated with naive splenocytes pulsed with 2 μg ml−1 OVA323–339 control or mITGB1697–724 peptide for a flow-based multi-cytokine array. Representative data from one of two
independent experiments using pools of five tumours each are shown as average of technical triplicate wells from three pooled tumours. Source data EXTENDED DATA FIG. 4 T3 TIL-DERIVED CD4+ T
CELL HYBRIDOMAS ARE REACTIVE AGAINST MITGB1. CTLL assay of T3 TIL-derived CD4+ T cell hybridoma lines stimulated with naive splenocytes pulsed with 2 μg ml−1 of the individual indicated
peptides. Representative data from one of three independent experiments are shown as average cpm from technical duplicate wells. Source data EXTENDED DATA FIG. 5 THE MITGB1 EPITOPE IS
PRESENTED ON I-AB. A, T3 CD4+ T cell hybridomas were stimulated with 2 μg ml−1 mITGB1(710Y) or wild-type ITGB1(710N) peptide-pulsed splenocytes. Activation was measured by CTLL assay.
Representative data from three independent hybridoma lines are shown as average of technical replicate wells. B, Mapping of the mITGB1 MHC-II binding core was performed using the CD4+ T cell
hybridoma line 41 stimulated with naive splenocytes pulsed with 2 μg ml−1 of overlapping peptides covering mITGB1697–724. Red denotes the T3-specific mutant amino acid at position p1 of the
minimal epitope; underlining denotes the validated binding core. Green amino acids represent random residue substitutions used to specifically define valines at residues 715 and 718 as the
p6 and p9 MHC-II binding positions and the complete MHC-II binding core. Representative data from two independent experiments are shown as the average of technical triplicate wells. C,
MHC-II I-Ab staining of parental T3 cells, IFNγ-stimulated T3 cells and T3 cells transduced with a vector encoding CIITA (T3.CIITA). Representative data from one of three independent
experiments are shown. D, Mirror plot showing match between MS/MS spectra of the 14-mer peptide sequence encompassing the N710Y site of mITGB1 eluted from T3.CIITA cells (positive axis) and
a corresponding synthetic peptide (negative axis). Labelled _m_/_z_ values reflect those experimentally observed for the endogenous peptide, with peaks representing _b_ ions highlighted in
blue and _y_ ions in red. Source data EXTENDED DATA FIG. 6 MITGB1 CD4+ T CELLS ARE REQUIRED FOR TUMOUR REJECTION IN RESPONSE TO ICT. A, Comparison of total number of expressed missense
mutations between ten different MCA-induced sarcomas and KP9025 cells. Mutations were defined by whole-exome sequencing and RNA sequencing, and mutational load is shown on a per cell basis.
B, Comparison of predicted neoantigen MHC-I affinity values between KP9025 and MCA-induced sarcoma F244 for H-2Db (top) and H-2Kb (bottom). KP9025 cells were not predicted to express any
MHC-I neoantigens. C, _Rag2__−/−_ mice were subcutaneously injected with 1 × 106 KP.mLAMA4, KP.mITGB1, KP.mLAMA4.mITGB1 or KP.mSB2.SIINFEKL cells. Representative data from one of two
independent experiments are presented as tumour diameters from individual mice (_n_ = 5 mice per group for KP.mLAMA4, KP.mITGB1 and KP.mLAMA4.mITGB1 and _n_ = 3 mice for KP.mSB2.SIINFEKL
group per experiment). D, Wild-type syngeneic 129S4 mice were injected subcutaneously with 1 × 106 KP.mLAMA4, KP.mITGB1 or KP.mLAMA4.mITGB1 cells and treated with anti-PD-1 (top) or
anti-CTLA single agent ICT (bottom) on days 3, 6, and 9 after transplantation. Representative data from one of three independent experiments are shown as tumour diameters from individual
mice (_n_ = 5 in all groups per experiment). E, Survival curves for experiments in D and Fig. 2e (_n_ = 15 in all groups). Source data EXTENDED DATA FIG. 7 OUTGROWTH OF NONIMMUNOGENIC
SARCOMA CELLS EXPRESSING MHC-I NEOANTIGENS IS NOT A RESULT OF CANCER IMMUNOEDITING. A, _Rag2__−/−_ or wild-type 129S4 mice were injected with 1 × 106 KP9025 or KP.mLAMA4 cells and treated
with anti-PD-1, anti-CTLA or anti-PD-1 + anti-CTLA4 on days 3, 6 and 9 after injection. Tumours were removed once they reached a maximum diameter of 20 mm in any direction and sarcoma cell
lines were established ex vivo. Cell lines were stimulated with IFNγ to upregulate MHC-I and subsequently used to stimulate the mLAMA4-specific CD8+ 74.14 T cell clone. Secretion of IFNγ by
T cells was measured using enzyme-linked immunosorbent assay (ELISA). Representative data from two independent experiments are represented as the average of two independent tumour samples in
each group. B, Wild-type 129S4 mice were injected with 1 × 106 KP.mSB2.SIINFEKL cells and treated with anti-PD-1 + anti-CTLA4 combination ICT on days 3, 6 and 9 after injection. Tumours
were removed as in A. Established ex vivo cell lines were cloned by limiting dilution and parental KP.mSB2.SIINFEKL cells or individual clones from outgrown tumours were used to stimulate
the mSB2-specific C3 CD8+ T cell clone; production of IFNγ was quantified using ELISA. Representative data from four independent experiments are presented as average IFNγ concentration of
eight individual clones ± s.e.m. Significance was determine using an unpaired, two-tailed _t_-test. C, Cell surface staining of SIINFEKL-H-2-Kb expressed by unstimulated or IFNγ-stimulated
parental KP.mSB2.SIINFEKL cells or individual clones described in B. A representative histogram is shown. D, Quantification of mean ± s.e.m. SIINFEKL-H-2-Kb mean fluorescence intensity (MFI)
from eight individual clones in C. NS, not significant. E, Survival curves of wild-type 129S4 mice injected subcutaneously with 1 × 106 KP.mSB2.SIINFEKL.mITGB1 cells. Mice were treated with
control monoclonal antibodies or anti-PD-1 + anti-CTLA4 combination ICT on days 3, 6 and 9 after injection. _n_ = 10 mice per group from two independent experiments. ****_P_ = 1.5 × 10−5,
Mantel–Cox test. Source data EXTENDED DATA FIG. 8 MITGB1-SPECIFIC CD4+ T CELLS DISPLAY AN ACTIVATED TH1 PHENOTYPE. A, Whole TILs from KP.mLAMA4.mITGB1 tumours 12 days after transplantation
were stained with mITGB1-I-Ab tetramers. Populations were previously gated on viable CD11b−CD4+ cells. Representative data from one of two independent experiments with five pooled tumours
each are shown. B, mITGB1-I-Ab tetramer-negative and tetramer-positive cells described in A were analysed for expression of T-BET and FOXP3. Representative plots are shown. C, Quantification
of two independent experiments in B as average per cent of tetramer-negative and tetramer-positive cells staining positive for the indicated protein. Tumour-bearing mice were treated with
control monoclonal antibodies or anti-CTLA4 on days 3, 6, and 9 after transplantation where indicated. D, mITGB1-I-Ab tetramer-positive and tetramer-negative cells in A were analysed for
expression of PD-1. Representative plots are shown. E, Quantification of two independent experiments described in D shown as average per cent of tetramer-negative and tetramer-positive cells
staining positive for PD-1. F, mITGB1-I-Ab tetramer-positive cells in A were analysed for expression of the indicated proteins. Representative histograms from one of two independent
experiments using pools of five tumours each are shown. Source data EXTENDED DATA FIG. 9 CD4+ T CELL HELP IS REQUIRED AT THE TUMOUR SITE DURING PRIMARY AND MEMORY RESPONSES. A, _Rag2__−/−_
mice were simultaneously injected with 1 × 106 KP.mLAMA4 and KP.mLAMA4.mITGB1 cells on opposite flanks. Representative data from one of two independent experiments are shown as individual
tumour diameter (_n_ = 3 in each experiment). B, Wild-type 129S4 mice were injected with 1 × 106 KP.mITGB1 cells and were treated with anti-PD-1 + anti-CTLA4 combination ICT on days 3, 6,
and 9 after injection. Representative data from one of two individual experiments are shown as individual tumour diameters (_n_ = 5 in all experiments). C, Wild-type 129S4 mice were
simultaneously injected with 1 × 106 KP.mLAMA4 and KP.mLAMA4.mITGB1 cells on opposite flanks and treated as in B. Representative data from one of two individual experiments are shown as
individual tumour diameters (_n_ = 5 in all experiments). D, Wild-type 129S6 mice were injected subcutaneously with 2 × 106 T3 sarcoma cells and were treated with anti-PD-1 + anti-CTLA4
combination ICT on days 3, 6, and 9 after injection. Following tumour rejection and a 30-day recovery period, tumour-experienced mice were rechallenged with 2 × 106 T3 cells in the presence
of control monoclonal antibody or CD4-depleting antibody, or with irrelevant sarcoma cells. Representative data from one of two independent experiments are shown as average tumour diameter ±
s.e.m. (_n_ = 5 in all groups per experiment). E, Wild-type 129S4 mice were injected subcutaneously with 1 × 106 KP.mLAMA4.mITGB1 cells followed by surgical resection 10 days after
transplantation. After a 30-day recovery period, tumour-experienced mice were rechallenged with 1 × 106 KP9025, KP.mLAMA4.mITGB1, or KP.mLAMA4 cells. Representative data from one of two
independent experiments are shown as average tumour diameter ± s.e.m. (_n_ = 5 in all groups per experiment). ****_P_ = 2 × 10−6 by two-way ANOVA with multiple comparisons and Bonferroni
correction. F, Quantification of data from three independent experiments in Fig. 5c is shown as average number of spots ± s.e.m. (left) and average number of mITGB1-specific CD4+ cells ±
s.e.m. (right). **_P_ = 0.003, ****_P_ = 7.2 × 10−5 (unpaired, two-tailed _t_-test). G, CD45+Ly6G−MHCII+CD64+CD25−CD11b+F4/80+ macrophages in TILs from mice bearing the indicated
contralateral tumours were analysed for expression of iNOS 11 days after tumour transplant. Representative data from four independent experiments are shown. H, Quantification of iNOS+
macrophages from experiments in F as a per cent of total CD45+ cells. Data are shown as average ± s.e.m. of four independent experiments. *_P_ = 0.03 by unpaired, two-tailed _t_-test. I,
CD45+Ly6G−MHCII+CD64+CD25−CD11b+F4/80+ macrophages from the indicated contralateral tumours described were isolated 11 days after transplantation and analysed for expression of iNOS.
Representative plots from two independent experiments are shown. J, Quantification of iNOS+ macrophages from two independent experiments in H is shown as average per cent of total CD45+
cells. Source data EXTENDED DATA FIG. 10 GATING STRATEGIES FOR MULTI-COLOUR FLOW CYTOMETRY. Gating strategies for multi-colour flow cytometry analysis of tumour-infiltrating macrophage (A)
and T cell (B) populations. SUPPLEMENTARY INFORMATION 41586_2019_1671_MOESM1_ESM.PDF Supplementary Table 1 SUPPLEMENTARY TABLE 1: CHARACTERISTICS OF THE SCREENED PREDICTED MHC CLASS II
NEOANTIGENS. Sequences of mutant peptides identified in T3 sarcomas used in the screening experiments REPORTING SUMMARY 41586_2019_1671_MOESM3_ESM.PDF Supplementary Table 2 SUPPLEMENTARY
TABLE 2: EXPRESSED MUTATIONS IN THE KP9025 SARCOMA CELL LINE. The four non-synonymous point mutations expressed in the KP9025 sarcoma cell line are shown as the corresponding amino acid
substitution 41586_2019_1671_MOESM4_ESM.XLSX Supplementary Data 1 SUPPLEMENTARY DATA 1: T3 NUCLEOTIDE VARIANT CALLS. This file contains all the single nucleotide variants determined to
generate missense mutations for the methylcholanthrene (MCA)-induced T3 sarcoma cell line as detected by cDNA capture sequencing 41586_2019_1671_MOESM5_ESM.XLS Supplementary Data 2
SUPPLEMENTARY DATA 2: KP9025 NUCLEOTIDE VARIANT CALLS. This file contains all the single nucleotide variants determined to generate missense mutations for the oncogene driven KP9025 sarcoma
cell line as detected by cDNA capture sequencing SOURCE DATA SOURCE DATA FIG. 1 SOURCE DATA FIG. 2 SOURCE DATA FIG. 3 SOURCE DATA FIG. 4 SOURCE DATA FIG. 5 SOURCE DATA EXTENDED DATA FIG. 1
SOURCE DATA EXTENDED DATA FIG. 2 SOURCE DATA EXTENDED DATA FIG. 3 SOURCE DATA EXTENDED DATA FIG. 4 SOURCE DATA EXTENDED DATA FIG. 5 SOURCE DATA EXTENDED DATA FIG. 6 SOURCE DATA EXTENDED DATA
FIG. 7 SOURCE DATA EXTENDED DATA FIG. 8 SOURCE DATA EXTENDED DATA FIG. 9 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Alspach, E., Lussier, D.M.,
Miceli, A.P. _et al._ MHC-II neoantigens shape tumour immunity and response to immunotherapy. _Nature_ 574, 696–701 (2019). https://doi.org/10.1038/s41586-019-1671-8 Download citation *
Received: 13 February 2019 * Accepted: 12 September 2019 * Published: 23 October 2019 * Issue Date: 31 October 2019 * DOI: https://doi.org/10.1038/s41586-019-1671-8 SHARE THIS ARTICLE Anyone
you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by
the Springer Nature SharedIt content-sharing initiative