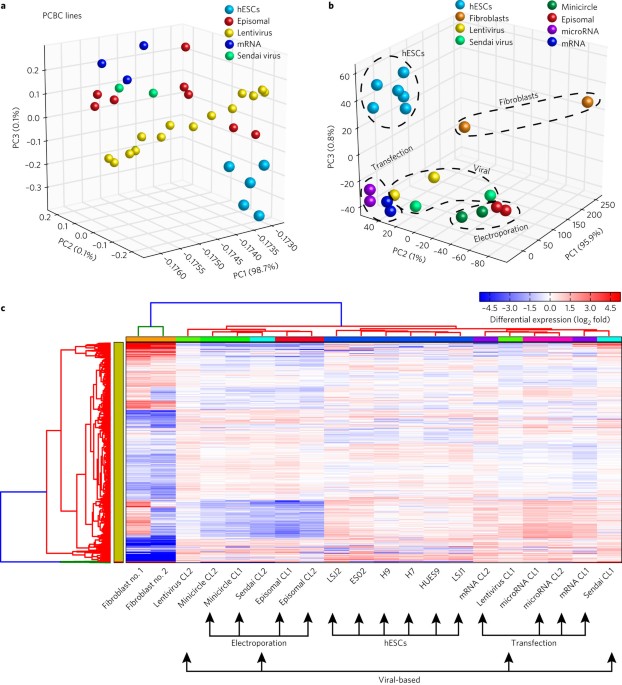
Transcriptomic and epigenomic differences in human induced pluripotent stem cells generated from six reprogramming methods
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Many reprogramming methods can generate human induced pluripotent stem cells (hiPSCs) that closely resemble human embryonic stem cells (hESCs). This has led to assessments of how
similar hiPSCs are to hESCs, by evaluating differences in gene expression, epigenetic marks and differentiation potential. However, all previous studies were performed using hiPSCs acquired
from different laboratories, passage numbers, culturing conditions, genetic backgrounds and reprogramming methods, all of which may contribute to the reported differences. Here, by using
high-throughput sequencing under standardized cell culturing conditions and passage number, we compare the epigenetic signatures (H3K4me3, H3K27me3 and HDAC2 ChIP-seq profiles) and
transcriptome differences (by RNA-seq) of hiPSCs generated from the same primary fibroblast population by using six different reprogramming methods. We found that the reprogramming method
impacts the resulting transcriptome and that all hiPSC lines could terminally differentiate, regardless of the reprogramming method. Moreover, by comparing the differences between the hiPSC
and hESC lines, we observed a significant proportion of differentially expressed genes that could be attributed to polycomb repressive complex targets. Access through your institution Buy or
subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get
Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 digital issues and online access to articles $119.00 per
year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during
checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS HIGH-RESOLUTION
SINGLE-CELL RNA-SEQ DATA AND HETEROGENEITY ANALYSIS OF HUMAN ESCS AND FFEPSCS Article Open access 22 April 2025 LONGITUDINAL ANALYSIS OF GENETIC AND EPIGENETIC CHANGES IN HUMAN PLURIPOTENT
STEM CELLS IN THE LANDSCAPE OF CULTURE-INDUCED ABNORMALITY Article Open access 01 November 2024 COMPLEX REGULATORY NETWORKS INFLUENCE PLURIPOTENT CELL STATE TRANSITIONS IN HUMAN IPSCS
Article Open access 23 February 2024 REFERENCES * Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors.
_Cell_ 126, 663–676 (2006). Article CAS PubMed Google Scholar * Chin, M. H. et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression
signatures. _Cell Stem Cell_ 5, 111–123 (2009). Article CAS PubMed PubMed Central Google Scholar * Wernig, M. et al. In vitro reprogramming of fibroblasts into a pluripotent
ES-cell-like state. _Nature_ 448, 318–324 (2007). Article CAS PubMed Google Scholar * Bock, C. et al. Reference maps of human ES and iPS cell variation enable high-throughput
characterization of pluripotent cell lines. _Cell_ 144, 439–452 (2011). Article CAS PubMed PubMed Central Google Scholar * Choi, J. et al. A comparison of genetically matched cell lines
reveals the equivalence of human iPSCs and ESCs. _Nat. Biotechnol._ 33, 1173–1181 (2015). Article CAS PubMed PubMed Central Google Scholar * Ruiz, S. et al. Identification of a
specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. _Proc. Natl Acad. Sci. USA_ 109, 16196–16201 (2012). Article CAS PubMed PubMed Central
Google Scholar * Newman, A. M. & Cooper, J. B. Lab-specific gene expression signatures in pluripotent stem cells. _Cell Stem Cell_ 7, 258–262 (2010). Article CAS PubMed Google
Scholar * Guenther, M. G. et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. _Cell Stem Cell_ 7, 249–257 (2010). Article CAS
PubMed PubMed Central Google Scholar * Wang, Y. et al. A transcriptional roadmap to the induction of pluripotency in somatic cells. _Stem Cell Rev._ 6, 282–296 (2010). Article CAS
PubMed Google Scholar * Kim, K. et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. _Nat. Biotechnol._ 29, 1117–1119
(2011). Article CAS PubMed PubMed Central Google Scholar * Fusaki, N., Ban, H., Nishiyama, A., Saeki, K. & Hasegawa, M. Efficient induction of transgene-free human pluripotent stem
cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. _Proc. Jpn Acad. Ser. B Phys. Biol. Sci._ 85, 348–362 (2009). Article CAS PubMed
PubMed Central Google Scholar * Gifford, C. A. et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. _Cell_ 153, 1149–1163 (2013). Article CAS
PubMed PubMed Central Google Scholar * Delgado-Olguin, P. et al. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. _Nat.
Genet._ 44, 343–347 (2012). Article CAS PubMed PubMed Central Google Scholar * Kyttala, A. et al. Genetic variability overrides the impact of parental cell type and determines iPSC
differentiation potential. _Stem Cell Rep._ 6, 200–212 (2016). Article Google Scholar * Bhutani, K. et al. Whole-genome mutational burden analysis of three pluripotency induction methods.
_Nat. Commun._ 7, 10536 (2016). Article CAS PubMed PubMed Central Google Scholar * Rouhani, F. et al. Genetic background drives transcriptional variation in human induced pluripotent
stem cells. _PLoS Genet._ 10, e1004432 (2014). Article PubMed PubMed Central Google Scholar * Heilig, C. et al. Implications of glucose transporter protein type 1 (GLUT1)-haplodeficiency
in embryonic stem cells for their survival in response to hypoxic stress. _Am. J. Pathol._ 163, 1873–1885 (2003). Article CAS PubMed PubMed Central Google Scholar * Janaszak-Jasiecka,
A. et al. miR-429 regulates the transition between hypoxia-inducible factor (HIF)1A and HIF3A expression in human endothelial cells. _Sci. Rep._ 6, 22775 (2016). Article CAS PubMed PubMed
Central Google Scholar * Wang, C. et al. Hypoxia inhibits myogenic differentiation through p53 protein-dependent induction of Bhlhe40 protein. _J. Biol. Chem._ 290, 29707–29716 (2015).
Article CAS PubMed PubMed Central Google Scholar * Bhandari, D. R. et al. The regulatory role of c-MYC on HDAC2 and PcG expression in human multipotent stem cells. _J. Cell. Mol. Med._
15, 1603–1614 (2011). Article CAS PubMed PubMed Central Google Scholar * Marshall, G. M. et al. Transcriptional upregulation of histone deacetylase 2 promotes Myc-induced oncogenic
effects. _Oncogene_ 29, 5957–5968 (2010). Article CAS PubMed Google Scholar * Zhang, Z. & Wu, W. S. Sodium butyrate promotes generation of human induced pluripotent stem cells
through induction of the miR302/367 cluster. _Stem Cells Dev._ 22, 2268–2277 (2013). Article CAS PubMed PubMed Central Google Scholar * Huangfu, D. et al. Induction of pluripotent stem
cells by defined factors is greatly improved by small-molecule compounds. _Nat. Biotechnol._ 26, 795–797 (2008). Article CAS PubMed Google Scholar * Kim, K. et al. Epigenetic memory in
induced pluripotent stem cells. _Nature_ 467, 285–290 (2010). Article CAS PubMed PubMed Central Google Scholar * Okita, K. et al. A more efficient method to generate integration-free
human iPS cells. _Nat. Methods_ 8, 409–412 (2011). Article CAS PubMed Google Scholar * Narsinh, K. H. et al. Generation of adult human induced pluripotent stem cells using nonviral
minicircle DNA vectors. _Nat. Protoc._ 6, 78–88 (2011). Article CAS PubMed Google Scholar * Warren, L. et al. Highly efficient reprogramming to pluripotency and directed differentiation
of human cells with synthetic modified mRNA. _Cell Stem Cell_ 7, 618–630 (2010). Article CAS PubMed PubMed Central Google Scholar * Anokye-Danso, F. et al. Highly efficient
miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. _Cell Stem Cell_ 8, 376–388 (2011). Article CAS PubMed PubMed Central Google Scholar * Liao, B. et al.
MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. _J. Biol. Chem._ 286, 17359–17364 (2011). Article CAS PubMed PubMed
Central Google Scholar * Sharma, A. et al. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. _J. Biol. Chem._ 288, 18439–18447 (2013). Article
CAS PubMed PubMed Central Google Scholar * Warlich, E. et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. _Mol. Ther._ 19,
782–789 (2011). Article CAS PubMed PubMed Central Google Scholar * Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features.
_Bioinformatics_ 26, 841–842 (2010). Article CAS PubMed PubMed Central Google Scholar * Krzywinski, M. I. et al. Circos: An information aesthetic for comparative genomics. _Genome Res_.
19, 1639–1645 (2009). * Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). _Genome Biol._ 9, R137 (2008). Article PubMed PubMed Central Google Scholar * McLean, C. Y. et al.
GREAT improves functional interpretation of _cis_-regulatory regions. _Nat. Biotechnol._ 28, 495–501 (2010). Article CAS PubMed PubMed Central Google Scholar * Sun, N. et al.
Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. _Sci. Transl. Med._ 4, 130ra147 (2012). Article Google Scholar * Huber, B. C. et al.
Costimulation-adhesion blockade is superior to cyclosporine A and prednisone immunosuppressive therapy for preventing rejection of differentiated human embryonic stem cells following
transplantation. _Stem Cells_ 31, 2354–2363 (2013). Article CAS PubMed PubMed Central Google Scholar * Emig, D. et al. AltAnalyze and DomainGraph: analyzing and visualizing exon
expression data. _Nucleic Acids Res._ 38, W755–W762 (2010). Article CAS PubMed PubMed Central Google Scholar * Kasprzyk, A. et al. EnsMart: a generic system for fast and flexible access
to biological data. _Genome Res._ 14, 160–169 (2004). Article CAS PubMed PubMed Central Google Scholar * Trapnell, C. et al. Differential gene and transcript expression analysis of
RNA-seq experiments with TopHat and Cufflinks. _Nat. Protoc._ 7, 562–578 (2012). Article CAS PubMed PubMed Central Google Scholar * Chen, J., Bardes, E. E., Aronow, B. J. & Jegga,
A. G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. _Nucleic Acids Res._ 37, W305–W311 (2009). Article CAS PubMed PubMed Central Google Scholar *
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. _Proc. Natl Acad. Sci. USA_ 102, 15545–15550 (2005). Article
CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This study was funded by the Canadian Institute of Health Research 201210MFE-289547 (J.M.C.), National
Institutes of Health 1K99HL128906 (J.M.C.), PCBC_JS_2014/4_01 (J.M.C.), National Research Foundation of Korea 2012R1A6A3A03039821 (J.L.), the Burroughs Wellcome Foundation, National
Institutes of Health R01 HL123968, HL128170, R01 HL126527 (J.C.W.), and P01 GM099130 (M.P.S.). The authors would like to thank the Stanford Stem Cell Institute Genome Center for their
sequencing knowledge, V. Sebastiano for hESC culturing, and B. Huber for his help with the teratoma assay. We would also like to thank J. Brito and B. Wu for their help in editing the
manuscript. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Institute of Stem Cell Biology and Regenerative Medicine, Stanford University School of Medicine, Stanford, CA, 94305, USA Jared M.
Churko, Jaecheol Lee, Mohamed Ameen, Mingxia Gu, Sebastian Diecke, Karim Sallam, Joseph D. Gold & Joseph C. Wu * Stanford Cardiovascular Institute, Stanford University School of
Medicine, Stanford, CA, 94305, USA Jared M. Churko, Jaecheol Lee, Mohamed Ameen, Mingxia Gu, Sebastian Diecke, Karim Sallam & Joseph C. Wu * Department of Medicine, Stanford University
School of Medicine, Stanford, CA, 94305, USA Jared M. Churko, Jaecheol Lee, Mohamed Ameen, Mingxia Gu, Sebastian Diecke, Karim Sallam & Joseph C. Wu * Division of Biomedical Informatics,
Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, 45229, USA Meenakshi Venkatasubramanian, Hogune Im & Nathan Salomonis * Department of Genetics, Stanford University School
of Medicine, Stanford, CA, 94305, USA Gavin Wang & Michael P. Snyder Authors * Jared M. Churko View author publications You can also search for this author inPubMed Google Scholar *
Jaecheol Lee View author publications You can also search for this author inPubMed Google Scholar * Mohamed Ameen View author publications You can also search for this author inPubMed Google
Scholar * Mingxia Gu View author publications You can also search for this author inPubMed Google Scholar * Meenakshi Venkatasubramanian View author publications You can also search for
this author inPubMed Google Scholar * Sebastian Diecke View author publications You can also search for this author inPubMed Google Scholar * Karim Sallam View author publications You can
also search for this author inPubMed Google Scholar * Hogune Im View author publications You can also search for this author inPubMed Google Scholar * Gavin Wang View author publications You
can also search for this author inPubMed Google Scholar * Joseph D. Gold View author publications You can also search for this author inPubMed Google Scholar * Nathan Salomonis View author
publications You can also search for this author inPubMed Google Scholar * Michael P. Snyder View author publications You can also search for this author inPubMed Google Scholar * Joseph C.
Wu View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS J.D.G., N.S., M.P.S. and J.C.W. supervised and planned the project. J.M.C. wrote the
manuscript, performed data analysis, generated and cultured hiPSC lines, and performed RNA-seq. N.S. and M.V. performed integration analysis. H.I. helped analyse RNA-seq. J.L. performed
ChIP-seq experiments. M.A. and M.G. performed FACS analysis on differentiated cardiomyocytes. G.W. and K.S. helped to culture hiPSC and hESC lines. S.D. generated minicircle hiPSC lines.
CORRESPONDING AUTHOR Correspondence to Joseph C. Wu. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE:
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY INFORMATION
Supplementary figures LIFE SCIENCES REPORTING SUMMARY SUPPLEMENTARY DATASET 1 Reads per kilobase of transcript per million mapped reads of each Ensembl ID, calculated via AltAnalyze.
SUPPLEMENTARY DATASET 2 Gene-expression differences, calculated via a Bayes moderated t-test p-value (unpaired), assuming unequal variance, and _p_ < 0.05 with two-fold difference.
SUPPLEMENTARY DATASET 3 Splicing events between hiPSCs and hESCs. SUPPLEMENTARY DATASET 4 Differential peaks unique to the hESCs and peaks unique to hiPSCs, corresponding to HDAC2
localization. SUPPLEMENTARY DATASET 5 Transcriptional-start-site peak-density differences within the H3K4me3 ChIP-seq set between hESCs and hiPSCs. SUPPLEMENTARY DATASET 6
Transcriptional-start-site peak-density differences in the H3K27me3 ChIP-seq profile between hESCs and hiPSCs. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Churko, J.M., Lee, J., Ameen, M. _et al._ Transcriptomic and epigenomic differences in human induced pluripotent stem cells generated from six reprogramming methods. _Nat Biomed Eng_
1, 826–837 (2017). https://doi.org/10.1038/s41551-017-0141-6 Download citation * Received: 24 October 2016 * Accepted: 04 September 2017 * Published: 03 October 2017 * Issue Date: October
2017 * DOI: https://doi.org/10.1038/s41551-017-0141-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative