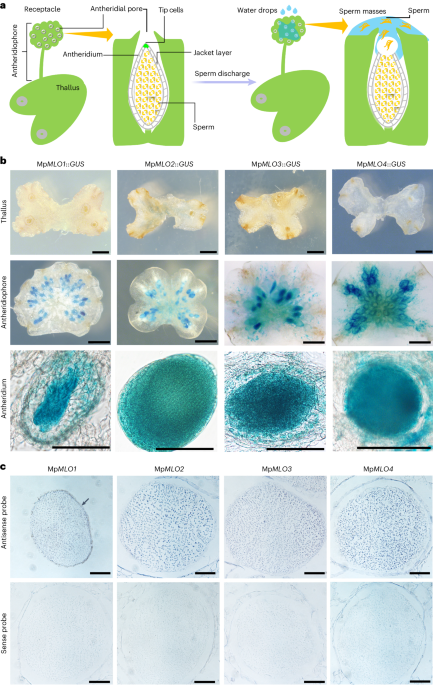
Mpmlo1 controls sperm discharge in liverwort
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT In bryophytes, sexual reproduction necessitates the release of motile sperm cells from a gametophyte into the environment. Since 1856, this process, particularly in liverworts, has
been known to depend on water. However, the molecular mechanism underlying this phenomenon has remained elusive. Here we identify the plasma membrane protein MpMLO1 in _Marchantia
polymorpha_, a model liverwort, as critical for sperm discharge from antheridia. The Mp_MLO1_-expressing tip cells among the sperm-wrapping jacket cells undergo programmed cell death upon
antheridium maturation to facilitate sperm discharge after the application of water and even hypertonic solutions. The absence of Mp_MLO1_ leads to reduced cytoplasmic Ca2+ levels in tip
cells, preventing cell death and consequently sperm discharge. Our findings reveal that MpMLO1-mediated programmed cell death in antheridial tip cells, regulated by cytosolic Ca2+ dynamics,
is essential for sperm release, elucidating a key mechanism in bryophyte sexual reproduction and providing insights into terrestrial plant evolution. Access through your institution Buy or
subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get
Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 digital issues and online access to articles $119.00 per
year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during
checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS SYP72 INTERACTS WITH
THE MECHANOSENSITIVE CHANNEL MSL8 TO PROTECT POLLEN FROM HYPOOSMOTIC SHOCK DURING HYDRATION Article Open access 10 January 2022 A NON-CANONICAL BZR/BES TRANSCRIPTION FACTOR REGULATES THE
DEVELOPMENT OF HAPLOID REPRODUCTIVE ORGANS IN _MARCHANTIA POLYMORPHA_ Article 11 April 2024 FLUORESCENCE SHADOW IMAGING OF _HYPSIBIUS EXEMPLARIS_ REVEALS MORPHOLOGICAL DIFFERENCES BETWEEN
SUCROSE- AND CACL2-INDUCED OSMOBIOTES Article Open access 23 May 2024 DATA AVAILABILITY The data for the current study are available within the paper and the Supplementary Information or
from the corresponding author upon request. Source data are provided with this paper. REFERENCES * Bowman, J. L. et al. Insights into land plant evolution garnered from the _Marchantia
polymorpha_ genome. _Cell_ 171, 287–304.e215 (2017). Article CAS PubMed Google Scholar * Kenrick, P. & Crane, P. R. The origin and early evolution of plants on land. _Nature_ 389,
33–39 (1997). Article CAS Google Scholar * Yamaoka, S. et al. Generative cell specification requires transcription factors evolutionarily conserved in land plants. _Curr. Biol._ 28,
479–486.e475 (2018). Article CAS PubMed Google Scholar * Koi, S. et al. An evolutionarily conserved plant RKD factor controls germ cell differentiation. _Curr. Biol._ 26, 1775–1781
(2016). Article CAS PubMed Google Scholar * Rövekamp, M., Bowman, J. L. & Grossniklaus, U. _Marchantia_ MpRKD regulates the gametophyte–sporophyte transition by keeping egg cells
quiescent in the absence of fertilization. _Curr. Biol._ 26, 1782–1789 (2016). Article PubMed Google Scholar * Ligrone, R., Duckett, J. G. & Renzaglia, K. S. Major transitions in the
evolution of early land plants: a bryological perspective. _Ann. Bot._ 109, 851–871 (2012). Article PubMed PubMed Central Google Scholar * Shimamura, M. _Marchantia polymorpha_:
taxonomy, phylogeny and morphology of a model system. _Plant Cell Physiol._ 57, 230–256 (2016). Article CAS PubMed Google Scholar * Kohchi, T., Yamato, K. T., Ishizaki, K., Yamaoka, S.
& Nishihama, R. Development and molecular genetics of _Marchantia polymorpha_. _Annu. Rev. Plant Biol._ 72, 677–702 (2021). Article CAS PubMed Google Scholar * Muggoch, H. &
Walton, J. On the dehiscence of the antheridium and the part played by surface tension in the dispersal of spermatocytes in Bryophyta. _Proc. R. Soc. Lond. B_ 861, 448–461 (1941). Google
Scholar * Thuret, G. Discharge of antherozoids in _Fegatella_. _Mem. Soc. Sci. Nat. Cherbourg_ 4, 216 (1856). Google Scholar * Paolillo, D. J. The swimming sperms of land plants.
_BioScience_ 31, 367–373 (1981). Article Google Scholar * Peirce, G. J. Forcible discharge of the antherozoids in _Asterella californica_. _Bull. Torrey Bot. Club_ 29, 374–382 (1902).
Article Google Scholar * Cavers, F. Explosive discharge of antherozoids in _Fegatella conica_. _Ann. Bot._ OS-17, 270–274 (1903). Article Google Scholar * Andersen, E. N. Discharge of
sperms in _Marchantia domingensis_. _Bot. Gaz._ 92, 66–84 (1931). Article Google Scholar * Paolillo, D. J. The release of sperms from the antheridia of _Polytrichum juniperinum_ Hedw. _New
Phytol._ 74, 287–293 (1975). Article Google Scholar * Paolillo, D. J. On the release of sperms in _Atrichum_. _Am. J. Bot._ 64, 81–85 (1977). Article Google Scholar * Kusch, S., Pesch,
L. & Panstruga, R. Comprehensive phylogenetic analysis sheds light on the diversity and origin of the MLO family of integral membrane proteins. _Genome Biol. Evol._ 8, 878–895 (2016).
Article PubMed PubMed Central Google Scholar * Büschges, R. et al. The barley Mlo gene: a novel control element of plant pathogen resistance. _Cell_ 88, 695–705 (1997). Article PubMed
Google Scholar * Consonni, C. et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. _Nat. Genet._ 38, 716–720 (2006). Article CAS PubMed Google
Scholar * Chen, Z. et al. Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in _Arabidopsis_ root thigmomorphogenesis. _Plant Cell_ 21, 1972–1991 (2009). Article
CAS PubMed PubMed Central Google Scholar * Bidzinski, P. et al. Physiological characterization and genetic modifiers of aberrant root thigmomorphogenesis in mutants of _Arabidopsis
thaliana_ MILDEW LOCUS O genes. _Plant Cell Environ._ 37, 2738–2753 (2014). Article CAS PubMed Google Scholar * Jacott, C. N., Charpentier, M., Murray, J. D. & Ridout, C. J. Mildew
Locus O facilitates colonization by arbuscular mycorrhizal fungi in angiosperms. _New Phytol._ 227, 343–351 (2020). Article CAS PubMed PubMed Central Google Scholar * Meng, J. G. et al.
Integration of ovular signals and exocytosis of a Ca2+ channel by MLOs in pollen tube guidance. _Nat. Plants_ 6, 143–153 (2020). Article CAS PubMed Google Scholar * Yi, J., An, S. &
An, G. Os_MLO12_, encoding seven transmembrane proteins, is involved with pollen hydration in rice. _Plant Reprod._ 27, 169–180 (2014). Article CAS PubMed Google Scholar * Kessler, S.
A. et al. Conserved molecular components for pollen tube reception and fungal invasion. _Science_ 330, 968–971 (2010). Article CAS PubMed Google Scholar * Jacott, C. N., Ridout, C. J.
& Murray, J. D. Unmasking Mildew Resistance Locus O. _Trends Plant Sci._ 26, 1006–1013 (2021). Article CAS PubMed Google Scholar * Kabbage, M., Kessens, R., Bartholomay, L. C. &
Williams, B. The life and death of a plant cell. _Annu. Rev. Plant Biol._ 68, 375–404 (2017). Article CAS PubMed Google Scholar * Reape, T. J. & McCabe, P. F. Apoptotic-like
programmed cell death in plants. _New Phytol._ 180, 13–26 (2008). Article CAS PubMed Google Scholar * Garcia, R. & McFeeley, J. C. Determining the cellular concentration and osmotic
potential of plant tissues. _Am. Biol. Teach._ 40, 119–120 (1978). Article Google Scholar * Micheli, F. Pectin methylesterases: cell wall enzymes with important roles in plant physiology.
_Trends Plant Sci._ 6, 414–419 (2001). Article CAS PubMed Google Scholar * Gao, Q. et al. A receptor-channel trio conducts Ca2+ signalling for pollen tube reception. _Nature_ 607,
534–539 (2022). Article CAS PubMed PubMed Central Google Scholar * Ren, H. et al. Calcium signaling in plant programmed cell death. _Cells_ 10, 1089 (2021). Article CAS PubMed PubMed
Central Google Scholar * Zhao, Y. et al. An expanded palette of genetically encoded Ca2+ indicators. _Science_ 333, 1888–1891 (2011). Article CAS PubMed PubMed Central Google Scholar
* Bi, G. et al. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. _Cell_ 184, 3528–3541.e3512 (2021). Article CAS PubMed Google Scholar * Ngo, Q.
A., Vogler, H., Lituiev, D. S., Nestorova, A. & Grossniklaus, U. A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. _Dev. Cell_ 29,
491–500 (2014). Article CAS PubMed Google Scholar * Bergdolt, E. _Untersuchungen über Marchantiaceen_ (Gustav Fischer, 1926). * Mecchia, M. A. et al. The single _Marchantia polymorpha_
FERONIA homolog reveals an ancestral role in regulating cellular expansion and integrity. _Development_ https://doi.org/10.1242/dev.200580 (2022). * Chiyoda, S., Ishizaki, K., Kataoka, H.,
Yamato, K. T. & Kohchi, T. Direct transformation of the liverwort _Marchantia polymorpha_ L. by particle bombardment using immature thalli developing from spores. _Plant Cell Rep._ 27,
1467–1473 (2008). Article CAS PubMed Google Scholar * Bienz, M. The PHD finger, a nuclear protein-interaction domain. _Trends Biochem. Sci._ 31, 35–40 (2006). Article CAS PubMed
Google Scholar * Kasahara, M. et al. An adenylyl cyclase with a phosphodiesterase domain in basal plants with a motile sperm system. _Sci. Rep._ 6, 39232 (2016). Article CAS PubMed
PubMed Central Google Scholar * Sugano, S. S. et al. Efficient CRISPR/Cas9-based genome editing and its application to conditional genetic analysis in _Marchantia polymorpha_. _PLoS ONE_
13, e0205117 (2018). Article PubMed PubMed Central Google Scholar * Ishizaki, K. et al. Development of gateway binary vector series with four different selection markers for the
liverwort _Marchantia polymorpha_. _PLoS ONE_ 10, e0138876 (2015). Article PubMed PubMed Central Google Scholar * Wu, H., Xie, D.-J., Tang, Z., Shi, D. & Yang, W.-C. PINOID regulates
floral organ development by modulating auxin transport and interacts with MADS16 in rice. _Plant Biotechnol. J._ 18, 1778–1795 (2020). Article CAS PubMed PubMed Central Google Scholar
* Ding, Y. et al. GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. _Plant Cell_
18, 815–830 (2006). Article CAS PubMed PubMed Central Google Scholar * Chen, Y.-H. et al. The central cell plays a critical role in pollen tube guidance in _Arabidopsis_. _Plant Cell_
19, 3563–3577 (2007). Article CAS PubMed PubMed Central Google Scholar * Herburger, K. & Holzinger, A. Aniline blue and Calcofluor white staining of callose and cellulose in the
streptophyte green algae _Zygnema_ and _Klebsormidium_. _Bio Protoc._ 6, e1969 (2016). Article PubMed Google Scholar * Schindelin, J. et al. Fiji: an open-source platform for
biological-image analysis. _Nat. Methods_ 9, 676–682 (2012). Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank E. Wang (CAS Center for Excellence in
Molecular Plant Science) and J. Zuo (Institute of Genetics and Developmental Biology, CAS) for the WT _M. polymorpha_ plants, W.-C. Yang for critical comments and supervision, and the public
technology service centre (Institute of Genetic and Developmental Biology, CAS) for assistance in confocal microscopy. This work was supported by the National Natural Science Foundation of
China (grant no. 32170343) and the CAS Project for Young Scientists in Basic Research (grant no. YSBR-078). AUTHOR INFORMATION Author notes * These authors contributed equally: Meng-Xing
Cao, Shi-Zhen Li. AUTHORS AND AFFILIATIONS * Key Laboratory of Seed Innovation, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China Meng-Xing Cao,
Shi-Zhen Li & Hong-Ju Li * Center for Molecular Agrobiology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China Meng-Xing Cao, Shi-Zhen Li &
Hong-Ju Li * University of Chinese Academy of Sciences, Beijing, China Hong-Ju Li Authors * Meng-Xing Cao View author publications You can also search for this author inPubMed Google
Scholar * Shi-Zhen Li View author publications You can also search for this author inPubMed Google Scholar * Hong-Ju Li View author publications You can also search for this author inPubMed
Google Scholar CONTRIBUTIONS M.-X.C. and S.-Z.L. performed the experiments and analysed the data. H.-J.L. conceived the study and supervised the project. M.-X.C. S.-Z.L. and H.-J.L. wrote
the paper. CORRESPONDING AUTHOR Correspondence to Hong-Ju Li. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature
Plants_ thanks Moritz Nowack and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 EXPRESSION PATTERN OF MP_MLO_ GENES. A. RT-PCR analysis of
Mp_MLO_ genes in the vegetative tissues, antheridiophore and archegoniophore. The control in archegoniophores and antheridiophores are represented by _PHD_ and _CAPE_, while _EF1α_ is used
as internal reference. B. Histochemical GUS assay of the _MpMLO_ genes in rhizoids. No GUS signal was detected in the rhizoids of plants carrying the Mp_MLO_ promoter-driven GUS construct.
Bar, 1 mm. Source data EXTENDED DATA FIG. 2 MUTATION OF EACH MP_MLO_ GENE GENERATED BY CRISPR/CAS9. The gene-editing regions of Mp_mlo2_, Mp_mlo3_ and Mp_mlo4_ are all located in the first
extron. Extrons are showed with black blocks. Red asterisks, deleted bases. Red letter, inserted base. EXTENDED DATA FIG. 3 THE THALLI AND ANTHERIDIA OF THE WILD TYPE AND EACH MP_MLO_
MUTANT. A–E. Morphological characteristics of thallus in WT and Mp_mlo_ mutants. Scale bar, 1 mm. F. Quantification of the antheridial pore numbers in the SEM images. Data are shown as the
mean ± s.e.m. _n_ = 11 and 10 antheridiophores for each genotype. Two-tailed Student’s _t_-test: _p_ = 0.5668. n.s. no statistical significance. G. Quantification of the antheridia number in
H and I. Data are shown as the mean ± s.e.m. _n_ = 14 and 16 antheridiophores for each genotype. Two-tailed Student’s _t_-test: _p_ = 0.4230. H and I. Transverse sections of mature
antheridiophore in WT (H) and Mp_mlo1_ (I). Arrows, antheridia. Scale bar, 500 μm. J and K. Number and location of antheridia in wild type and Mp_mlo1_. Vertical section of antheridiophores
in Wild type (J) and Mp_mlo1 #1_ (K). The growth locations of antheridia in Mp_mlo1 #1_ is consistent with WT. Asterisks, antheridia. Scale bar, 100 μm. EXTENDED DATA FIG. 4 SPERM DISCHARGE
ASSAY IN MP_MLO1_, MP_MLO2_, MP_MLO3_ AND MP_MLO4_ MUTANTS. A–F. Sperm discharge after application of water drops. Red arrows, released sperm masses. Scale bar, 1 mm. G. Thickened jacket
layer in the two Mp_mlo1_ mutant lines. White arrows indicate the jacket cell layer; Scale bar, 100 µm. H. Quantification of the width of different Mp_mlo1_ mutant lines. Data are shown as
the mean ± s.e.m. _n_ = 12, 11 and 12 antheridia for each line (from left to right). Significant differences were determined using one-way ANOVA, different letters indicate values with
statistically significant (p < 0.05; Tukey honest significant difference [HSD]) and non-significant (p > 0.05; Tukey HSD) differences, respectively. EXTENDED DATA FIG. 5 DEVELOPMENT OF
JACKET LAYER IN WILD TYPE AND MP_MLO_ MUTANTS. Morphological characteristics of antheridia isolated at immature and mature stages in Mp_mlo_ mutants. Scale bar, 20 μm. EXTENDED DATA FIG. 6
DEVELOPMENT OF ANTHERIDIA IN WILD TYPE, MP_MLO2_, MP_MLO3_ AND MP_MLO4._ Vertical section of antheridia at different stages in wild type and the mutants of Mp_mlo2_, Mp_mlo3_, and Mp_mlo4_.
Antheridia in Mp_mlo2_, Mp_mlo3_, and Mp_mlo4_ show no obvious difference with wild type during the developmental stages. Scale bar, 20 μm. EXTENDED DATA FIG. 7 EXPRESSION OF MPMLO1-CITRINE
IN ANTHERIDIA. A–C. MpMLO1-Citrine expressed in whole-mount antheridia. Arrow, jacket layer. Scale bar, 50 μm. D–G. MpMLO1-Citrine expressed in flagella of sperm. Nuclei were stained by
DAPI. BF, bright field. Scale bar, 5 μm. EXTENDED DATA FIG. 8 TUNEL AND FDA-PI STAINING OF ANTHERIDIA IN WILD TYPE AND MP_MLO1_ MUTANT. A. TUNEL assay for the WT and Mp_mlo1_ mutants. The
images of TUNEL staining are representative of 141 antheridia. Three independent experiments were performed. Positive control, Dnase I treatment prior to TUNEL staining; Negative control,
TUNEL staining performed without adding TdT enzyme. Details shown in Method part. Scale bar, 100 µm. B. FDA-PI staining of immature antheridia. In A and B, the white arrows indicate the
antheridial tip cells. Scale bar, 100 µm. For A and B, three independent experiments were performed. EXTENDED DATA FIG. 9 HISTOCHEMICAL STAINING OF ANTHERIDIA IN WILD TYPE AND MP_MLO1_
MUTANT. A. Aniline blue staining of antheridia in MpMLO1-Citrine-expressing WT and Mp_mlo1 #1_. Red, callose. B. Calcofluor White staining of antheridia in MpMLO1-Citrine-expressing WT and
Mp_mlo1 #1_. Magenta, cellulose. C. JIM7 immunostaining of antheridia in MpMLO1-Citrine-expressing WT and Mp_mlo1 #1_. Red, esterified pectin. Green, fluorescence of MpMLO1-Citrine. For each
staining, three independent experiments were performed. Arrows, tip cells. Scale bar, 50 μm. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Table 1. REPORTING SUMMARY
SUPPLEMENTARY VIDEO 1 Swimming WT sperm cells. SUPPLEMENTARY VIDEO 2 Swimming Mp_mlo1_ sperm cells. SUPPLEMENTARY VIDEO 3 3D imaging of MpMLO1–Citrine in the immature antheridia.
SUPPLEMENTARY VIDEO 4 3D imaging of MpMLO1–Citrine in the mature antheridia. SUPPLEMENTARY VIDEO 5 Time-lapse imaging of Ca2+ in the WT antheridium. SUPPLEMENTARY VIDEO 6 Time-lapse imaging
of Ca2+ in the Mp_mlo1_ #1 antheridium. SUPPLEMENTARY VIDEO 7 Time-lapse imaging of Ca2+ in the Mp_mlo1_ #2 antheridium. SOURCE DATA SOURCE DATA EXTENDED DATA FIG. 1 DNA gels 1–4. RIGHTS AND
PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other
rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Cao, MX., Li, SZ. & Li, HJ. MpMLO1 controls sperm discharge in liverwort. _Nat. Plants_ 10, 1027–1038 (2024).
https://doi.org/10.1038/s41477-024-01703-1 Download citation * Received: 27 June 2023 * Accepted: 18 April 2024 * Published: 03 June 2024 * Issue Date: June 2024 * DOI:
https://doi.org/10.1038/s41477-024-01703-1 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative