Legionella pneumophila evades host-autophagic clearance using phosphoribosyl-polyubiquitin chains
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Legionella pneumophila, the causative agent of Legionnaires’ disease, colonises host cells through effective avoidance of host defense mechanisms. In this commentary, we highlight two recent
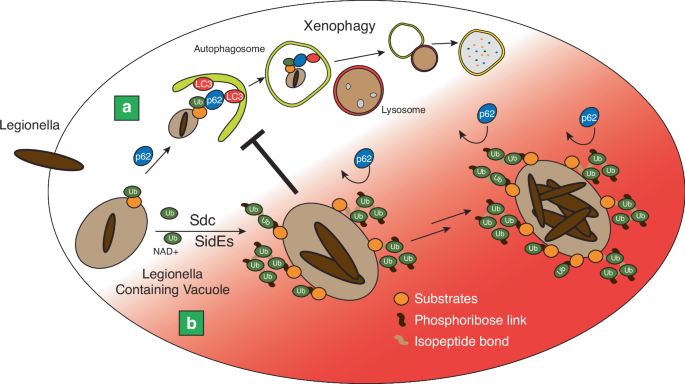
Nature Communications studies reporting that two Legionella effectors, Sdc and Sde, working together to decorate the Legionella-containing vacuole (LCV) with unusual mixed ubiquitin chains.
The phosphoribosyl modifications on the polyubiquitin chains block recognition by ubiquitin adaptors, thereby preventing the recruitment of host autophagy receptors like p62 to the LCV.
This allows Legionella to evade autophagic clearance and establish a replicative niche within host cells. LEGIONELLA PNEUMOPHILA CATALYZES UNUSUAL PHOSPHORIBOSYL-SERINE UBIQUITINATION The
SidE family of effector proteins (SidEs) from _Legionella pneumophila_ catalyze a novel form of ubiquitination independent of the canonical E1-E2-E3 enzyme cascade1,2. First, the
mono-ADP-ribosyltransferase (mART) domain of SidEs catalyzes the transfer of ADP-ribose from NAD+ to arginine 42 of ubiquitin, generating ADP-ribosylated ubiquitin (ADPR-Ub). The
phosphodiesterase (PDE) domain then processes ADPR-Ub to phosphoribosylated ubiquitin (PR-Ub) by cleaving the pyrophosphate bond and releasing AMP. The PDE domain then catalyzes the
conjugation of PR-Ub to serine residues in substrate proteins via a phosphodiester bond. This occurs through a two-step mechanism: first, PR-Ub is linked to a catalytic histidine in the PDE
domain via a phosphoamidate linkage, forming a covalent intermediate. Secondly, the phosphoribosyl-ubiquitin is transferred from the catalytic histidine to a serine hydroxyl in the substrate
protein3,4,5. The activity of the SidE effectors is tightly regulated by other Legionella effectors. SidJ, a metaeffector, inhibits SidE-mediated PR-ubiquitination by glutamylating the mART
domain catalytic residue E860, thereby blocking the initial step of ADPR-Ub generation. SidJ glutamylation activity requires the eukaryotic co-factor calmodulin and is activated by
decreased cytosolic Ca2+ levels6,7,8. Additionally, DupA and DupB, two Legionella effectors with PDE domains homologous to SidEs, act as specific deubiquitinases that reverse PR-serine
ubiquitination9,10. Despite the structural similarity, DupA/B exhibit higher affinity for PR-ubiquitinated substrates compared to SidEs. This enables DupA/B to specifically cleave the
phosphodiester bond between PR-Ub and serine residues, counteracting SidE activity. The balance between SidE-mediated PR-ubiquitination and DupA/B-mediated deubiquitination dynamically
regulates the extent and duration of this unique post-translational modification during Legionella infection. LEGIONELLA PNEUMOPHILA SIDC/SDCA ARE UBIQUITIN LIGASES THAT FACILITATE BACTERIAL
PHAGOSOMAL REMODELING Another major family of _L. pneumophila_ ubiquitin ligases is the Sdc proteins (SidC and SdcA). In contrast to the SidE family, the Sdc effectors catalyze conventional
lysine-targeted ubiquitination via a novel E3 ubiquitin ligase domain. Initially, SidC and SdcA were proposed to act as vesicle tethering factors that recruit ER-derived vesicles to the
LCV11. However, structural and biochemical studies have revealed that SidC and SdcA possess E3 ubiquitin ligase activity, defining a novel family of bacterial ubiquitin ligases12,13. SidC
and SdcA each contain an N-terminal SNL (SidC N-terminal Ligase) domain and a C-terminal PI4P-binding domain that anchors them to the LCV membrane. The crystal structure of the SNL domain
revealed a unique fold with no homology to known ubiquitin ligases. Intriguingly, the SNL domain contains a canonical catalytic triad consisting of a cysteine, histidine, and aspartate
residue, reminiscent of the active site found in cysteine proteases and deubiquitinases. During infection, SidC/SdcA-mediated ubiquitination is important for recruitment of ER proteins and
ubiquitin to the LCV. A SidC mutant lacking E3 ligase activity fails to complement these recruitment defects in a ΔsidC-sdcA strain. SidC and SdcA appear to ubiquitinate different
substrates, as they exhibit distinct preferences for ubiquitin-conjugating E2 enzymes. This functional diversification likely allows Legionella to maximize its manipulation of host
ubiquitination pathways. The discovery of the SidC/SdcA E3 ligases expands the repertoire of strategies used by Legionella to hijack host cells. By combining ubiquitin ligase activity with
phospholipid binding and vesicle tethering capabilities, these novel effectors dynamically reshape the phagosomal membrane to promote Legionella survival and replication. SIDES AND SDC
COOPERATE TO AVOID RECOGNITION BY HOST-AUTOPHAGIC ADAPTORS The recent studies by Kotewicz et al. and Wan et al. have provided novel insights into the intricate interplay between the SidE and
Sdc effectors in shaping the unique ubiquitin landscape on the LCV14,15 (Fig. 1). Through a combination of biochemical, cell biological, and structural approaches, these studies have
unveiled a remarkable cooperative action between these two effector families, leading to the decoration of the LCV with unusual mixed ubiquitin chains. Kotewicz et al. focused on dissecting
the role of the SidE effectors’ deubiquitinase (DUB) domain, which had previously been shown to preferentially cleave K63-linked polyubiquitin chains. Surprisingly, they found that the DUB
domain, rather than reducing polyubiquitination on the LCV, actually promotes the PR-ubiquitination of Rtn4 and the subsequent ER remodeling. Through a series of elegant experiments, they
demonstrated that the DUB domain’s activity is critical for generating monoubiquitin from K63-linked chains, which in turn serves as a substrate for the mART domain to generate ADPR-Ub. This
ADPR-Ub is then used by the PDE domain for PR-ubiquitination of Rtn4. These findings provide a novel mechanistic understanding of how the different domains within SidE effectors cooperate
to fine-tune the ubiquitin signals on the LCV. Wan et al., on the other hand, took a broader approach to investigate the crosstalk between the SidE and SdC effectors. Through a combination
of immunoprecipitation and mass spectrometry analyses, they discovered that multiple host proteins, including Rab small GTPases and ER proteins, are co-modified by both conventional
ubiquitination (mediated by SidC) and PR-ubiquitination (mediated by SidE). This co-modification was found to be crucial for the efficient recruitment of these host proteins to the LCV and
the subsequent remodeling of the ER. Notably, they demonstrated that SidC and SidE effectors work cooperatively to build mixed ubiquitin chains, where SidC extends polyubiquitin chains on
substrates initially modified with PR-Ub by SidE. This intricate collaboration between the two effector families highlights the sophisticated strategies employed by L. pneumophila to
fine-tune the host-pathogen interface. A key finding that emerged from both studies is the identification of a novel mechanism by which L. pneumophila evades autophagic recognition and
clearance. Through mass spectrometry analysis, Kotewicz et al. and Wan et al. discovered that SidE effectors specifically modify the R42 residue of ubiquitin within polyubiquitin chains on
the LCV. This modification is of particular significance, as the R42 residue is a critical determinant for ubiquitin recognition by autophagy adaptors such as p62. By modifying this residue,
SidE effectors sterically hinder the binding of p62 to ubiquitin, effectively preventing the recruitment of autophagy receptors to the LCV. This discovery provides a mechanistic explanation
for how L. pneumophila can maintain an extensively ubiquitinated vacuole while avoiding autophagic degradation. Another intriguing finding from Wan et al. is the ability of SidE effectors
to generate PR-Ub linkages within polyubiquitin chains, leading to the crosslinking of multiple substrates into ubiquitin-tethered complexes on the LCV. They demonstrated this phenomenon by
identifying complexes containing Rab1, Rab33b, and the ER protein LULL1. This additional layer of complexity in the ubiquitin architecture on the LCV further underscores the intricate
strategies employed by L. pneumophila to create a specialized replicative niche. Furthermore, both studies shed light on the role of additional players in the PR-ubiquitination process. Wan
et al. identified the involvement of the PR-Ub deubiquitinases DupA and DupB in converting ADPR-Ub to PR-Ub within polyubiquitin chains. This finding expands our understanding of the
enzymatic repertoire that L. pneumophila employs to fine-tune the ubiquitin signals on the LCV and highlights the dynamic nature of these modifications. PERSPECTIVES The findings by Kotewicz
et al. and Wan et al. have significantly advanced our understanding of the sophisticated strategies employed by L. pneumophila to manipulate the host ubiquitin system. These studies have
unveiled a remarkable level of cooperation and crosstalk between the SidE and SidC effector families, leading to the generation of distinctive mixed ubiquitin chains on the LCV. The
discovery that these effectors collaborate to decorate the LCV with PR-ubiquitin marks, thereby camouflaging the vacuole from autophagic recognition, represents a novel mechanism by which a
pathogen can subvert host defenses and ensure its survival and replication. The identification of this intricate ubiquitin manipulation strategy raises intriguing questions and opens up new
avenues for future research. A key area of interest is to understand the temporal and spatial regulation of SidE and SidC activities during the course of infection. Deciphering the
mechanisms that govern the coordinated action of these effectors will provide valuable insights into how L. pneumophila fine-tunes the host-pathogen interface at different stages of its
intracellular lifecycle. Additionally, structural studies aimed at elucidating the molecular basis of the cooperation between SidE and SidC effectors in the assembling of mixed ubiquitin
chains will be crucial for a deeper mechanistic understanding of this process. Another exciting direction for future research is the investigation of the potential interplay between the
PR-ubiquitination machinery and other L. pneumophila effectors that target the host ubiquitin system. The L. pneumophila genome encodes a vast array of effectors with diverse functions, and
it is likely that the SidE and SidC effectors work in concert with other effectors to orchestrate the complex host-pathogen interactions. For example, a very recent study highlights that
LnaB of L.pneumophila reverses the PR-ubiquitin to ADPR-ubiquitin through the addition of AMP to the PR-ubiquitin16. It will be interesting to investigate the PR-moieties on the
polyubiquitin chains created by cooperative reaction of Sdc and SidE are also accessible with LnaB. From a broader perspective, the discovery of PR-ubiquitination as a mechanism to evade
autophagy raises the intriguing possibility that similar strategies may be employed by other intracellular pathogens. It will be interesting to explore whether PR-ubiquitination or related
modifications are utilized by other bacteria to subvert host defenses. Comparative studies across different pathogenic species could shed light on the evolutionary conservation and
diversification of ubiquitin manipulation strategies. In conclusion, the studies by Kotewicz et al. and Wan et al. represent a significant leap forward in our understanding of how L.
pneumophila hijacks the host ubiquitin system to create a permissive replicative niche. The discovery of the cooperative action of SidE and SidC effectors in generating mixed ubiquitin
chains and the identification of PR-ubiquitination as a mechanism to evade autophagy have unveiled new paradigms in host-pathogen interactions. These findings underscore the importance of
considering the complex interplay between bacterial effectors and the potential for novel post-translational modifications in shaping the outcome of infections. As we continue to unravel the
intricate strategies employed by pathogens to subvert host defenses, we will not only gain a deeper understanding of the pathogenesis of infectious diseases but also gain new insights into
fundamental cellular processes. The exciting discoveries made by Kotewicz et al. and Wan et al. pave the way for future research that will further illuminate the molecular battles between
pathogens and their hosts, ultimately informing the development of new therapeutic strategies to combat infections. REFERENCES * Bhogaraju, S. et al. Phosphoribosylation of Ubiquitin
Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. _Cell_ 167, 1636–1649.e1613 (2016). Article CAS PubMed Google Scholar * Qiu, J. et al. Ubiquitination independent
of E1 and E2 enzymes by bacterial effectors. _Nature_ 533, 120–124 (2016). Article ADS CAS PubMed PubMed Central Google Scholar * Akturk, A. et al. Mechanism of
phosphoribosyl-ubiquitination mediated by a single Legionella effector. _Nature_ 557, 729–733 (2018). Article ADS CAS PubMed PubMed Central Google Scholar * Dong, Y. et al. Structural
basis of ubiquitin modification by the Legionella effector SdeA. _Nature_ 557, 674–678 (2018). Article ADS CAS PubMed Google Scholar * Kalayil, S. et al. Insights into catalysis and
function of phosphoribosyl-linked serine ubiquitination. _Nature_ 557, 734–738 (2018). Article ADS CAS PubMed PubMed Central Google Scholar * Bhogaraju, S. et al. Inhibition of
bacterial ubiquitin ligases by SidJ-calmodulin catalysed glutamylation. _Nature_ 572, 382–386 (2019). Article ADS CAS PubMed PubMed Central Google Scholar * Black, M. H. et al.
Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases. _Science_ 364, 787–792 (2019). Article ADS CAS PubMed PubMed Central Google
Scholar * Gan, N. et al. Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase. _Nature_ 572, 387–391 (2019). Article ADS CAS PubMed PubMed Central Google
Scholar * Shin, D. et al. Regulation of Phosphoribosyl-Linked Serine Ubiquitination by Deubiquitinases DupA and DupB. _Mol. Cell_ 77, 164–179.e166 (2020). Article CAS PubMed PubMed
Central Google Scholar * Wan, M. et al. Deubiquitination of phosphoribosyl-ubiquitin conjugates by phosphodiesterase-domain-containing Legionella effectors. _Proc. Natl Acad. Sci. USA_
116, 23518–23526 (2019). Article ADS CAS PubMed PubMed Central Google Scholar * Ragaz, C. et al. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate
SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. _Cell Microbiol_ 10, 2416–2433 (2008). Article CAS PubMed Google Scholar * Horenkamp, F. A. et al.
Legionella pneumophila subversion of host vesicular transport by SidC effector proteins. _Traffic_ 15, 488–499 (2014). Article CAS PubMed PubMed Central Google Scholar * Hsu, F. et al.
The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling. _Proc. Natl Acad. Sci. USA_ 111, 10538–10543 (2014). Article ADS
CAS PubMed PubMed Central Google Scholar * Kotewicz, K. M. et al. Sde proteins coordinate ubiquitin utilization and phosphoribosylation to establish and maintain the Legionella
replication vacuole. _Nat. Commun._ https://doi.org/10.1038/s41467-024-51272-2 (2024). * Wan, M. et al. Phosphoribosyl modification of poly-ubiquitin chains at the Legionella-containing
vacuole prohibiting autophagy adaptor recognition. _Nat. Commun._ https://doi.org/10.1038/s41467-024-51273-1 (2024). * Wang, T. et al. Legionella effector LnaB is a phosphoryl-AMPylase that
impairs phosphosignalling. _Nature_ https://doi.org/10.1038/s41586-024-07573-z (2024). Download references ACKNOWLEDGEMENTS Research in the D.S. laboratory is supported by a National
Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2021R1C1C100396112, 2018R1A6A1A0302560722 and 2021M3A9I4021220), the Yonsei University Research Fund of
2021 (2021-22-0050), Korea Disease Control and Prevention Agency Research Fund (2023-ER2002-01), Yuhan Innovation Program 2023 by Yuhan Corporation and POSCO Science Fellowship by POSCO TJ
Park Foundation. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Systems Biology, College of Life Science and Biotechnology, Yonsei University, 03722, Seoul, Republic of Korea
Minhyeong Choi, Minwoo Jeong, Sangwoo Kang, Hayoung Jeon & Donghyuk Shin Authors * Minhyeong Choi View author publications You can also search for this author inPubMed Google Scholar *
Minwoo Jeong View author publications You can also search for this author inPubMed Google Scholar * Sangwoo Kang View author publications You can also search for this author inPubMed Google
Scholar * Hayoung Jeon View author publications You can also search for this author inPubMed Google Scholar * Donghyuk Shin View author publications You can also search for this author
inPubMed Google Scholar CONTRIBUTIONS S.K., M.C., H.J. wrote the manuscript, M.J. drew the figure. D.S. designed overall structure of manuscript and wrote the manuscript. CORRESPONDING
AUTHOR Correspondence to Donghyuk Shin. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give
appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission
under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons
licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by
statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Choi, M., Jeong, M., Kang, S. _et al._ Legionella pneumophila evades
host-autophagic clearance using phosphoribosyl-polyubiquitin chains. _Nat Commun_ 15, 7480 (2024). https://doi.org/10.1038/s41467-024-51277-x Download citation * Received: 15 June 2024 *
Accepted: 02 August 2024 * Published: 30 August 2024 * DOI: https://doi.org/10.1038/s41467-024-51277-x SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative