A citric acid cycle-deficient escherichia coli as an efficient chassis for aerobic fermentations
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Tricarboxylic acid cycle (TCA cycle) plays an important role for aerobic growth of heterotrophic bacteria. Theoretically, eliminating TCA cycle would decrease carbon dissipation and
facilitate chemicals biosynthesis. Here, we construct an _E. coli_ strain without a functional TCA cycle that can serve as a versatile chassis for chemicals biosynthesis. We first use
adaptive laboratory evolution to recover aerobic growth in minimal medium of TCA cycle-deficient _E. coli_. Inactivation of succinate dehydrogenase is a key event in the evolutionary
trajectory. Supply of succinyl-CoA is identified as the growth limiting factor. By replacing endogenous succinyl-CoA dependent enzymes, we obtain an optimized TCA cycle-deficient _E. coli_
strain. As a proof of concept, the strain is engineered for high-yield production of four separate products. This work enhances our understanding of the role of the TCA cycle in _E. coli_
metabolism and demonstrates the advantages of using TCA cycle-deficient _E. coli_ strain for biotechnological applications. SIMILAR CONTENT BEING VIEWED BY OTHERS _ESCHERICHIA COLI_ IS
ENGINEERED TO GROW ON CO2 AND FORMIC ACID Article 28 September 2020 REPROGRAMMING METHANOL UTILIZATION PATHWAYS TO CONVERT _SACCHAROMYCES CEREVISIAE_ TO A SYNTHETIC METHYLOTROPH Article 15
May 2023 INCREASED CO2 FIXATION ENABLES HIGH CARBON-YIELD PRODUCTION OF 3-HYDROXYPROPIONIC ACID IN YEAST Article Open access 21 February 2024 INTRODUCTION Metabolic engineering of industrial
microbes to synthesize value-added products from renewable carbon sources requires high product yields and substrate utilization rates. Microbes have naturally evolved to convert available
substrates into cell mass; any attempts to reroute carbon fluxes away from cell growth and towards products of interest disturbs native metabolism and can affect carbon efficiency. For the
heterotrophic industrial bacteria _E. coli_, the tricarboxylic acid cycle (TCA cycle) plays multiple roles during aerobic growth. In addition to providing key precursors for biomass
synthesis, the conventional TCA cycle completely oxidizes carbon sources such as glucose to CO2, generates reductants (NAD(P)H, FADH2), and produces energy (ATP) primarily through coupling
with oxidative phosphorylation. The TCA cycle oxidizes each acetyl-CoA to two CO2. The emission of CO2 from the TCA cycle reduces carbon fluxes directed to products synthesis, thus
negatively impacting product yield. Therefore, blocking the TCA cycle and its bypass pathways could decrease carbon dissipation and facilitate chemical biosynthesis in aerobic fermentations.
Strategies that block or shunt TCA cycle flux to raise product yields have thus far encountered growth defects in minimal medium1,2,3. For example, inactivation of α-ketoglutarate
dehydrogenase (_ΔsucA_) and glyoxylate shunt (_ΔaceA_)1,2,3 resulted in no aerobic growth on glucose. Several strategies have been attempted to overcome this defect. One way was to use an
inducible switch of the metabolic flux at the α-ketoglutarate node. By introducing a genetic circuit based on a dual-induction system, conditional interruption of the TCA cycle was used to
increase the yield of γ-aminobutyric acid from glucose4. Another strategy was to bypass the α-ketoglutarate dehydrogenase reaction using α-ketoglutarate-dependent oxygenase, an enzyme that
converts α-ketoglutarate to succinate. As an example, growth of _E. coli ΔaceAΔsucA_ strain was enabled on glucose by introducing L-isoleucine dioxygenase1 or L-proline-4-hydroxylase
(P4H)2,3 with the additional supplementation of isoleucine or proline. However, this latter strategy has several limitations: the corresponding α-ketoglutarate-dependent oxygenases must be
active enough in _E. coli_, the co-substrates must be efficiently imported or synthesized, and the substrates and products must not be toxic to the cells1. Adaptive laboratory evolution
(ALE) is a strategy often used to overcome growth defects of engineered strains5,6,7. For example, ALE has been successfully applied to recover aerobic growth of various _E. coli_ mutant
strains: a strain without phosphotransferase system (PTS)8, a low NADPH-producing (_ΔzwfΔeddΔeda_) strain9, and a phosphoglucose isomerase (_pgi_) defective high NADPH-producing strain10,
just to mention a few. In ALE studies, comparative omics analyses and isotope-labeled metabolism flux analysis (13C-MFA)11 are standard procedures for dissecting the genetic basis and
metabolic mechanisms for growth recovery. A systematic analysis of an evolved TCA cycle-deficient _E. coli_ would be valuable for metabolic engineers who desire to attenuate TCA cycle fluxes
in favor of higher product yields. In previous work12, a whole-cell biocatalytic process was developed that employed an engineered TCA cycle-deficient (_ΔaceAΔsucA_) _E. coli_ strain
expressing α-ketoglutarate-dependent deacetoxycephalosporin C synthase (DAOCS) to convert penicillin G to G-7-ADCA. Initially, the TCA cycle-deficient strain was cultured with a rich medium
and induced for overexpression of deacetoxycephalosporin C synthase, without the presence of penicillin G, during the cell growth stage. Subsequently, the resting cells, which served as the
biocatalyst, were used to produce deacetoxycephalosporin C with penicillin G functioning as the substrate12. However, due to the toxic nature of both the substrate and products of DAOCS on
_E. coli_, it was impossible to repair the TCA cycle using the DAOCS reaction while cultivating this engineered strain in a glucose minimal medium. In this work, a TCA cycle-deficient _E.
coli_ is adaptively evolved to restore aerobic growth in glucose minimal medium. Whole genome sequencing and 13C-MFA analysis uncover the mechanisms for growth recovery and identify specific
metabolic bottlenecks that are subsequently resolved by targeted engineering of metabolic pathways. We thus construct an efficient _E. coli_ strain without a TCA cycle that can be used for
production of chemicals. As a proof of concept, we use the optimized _E. coli_ strain for high-yield biosynthesis of four separate products in aerobic fermentations, thus demonstrating the
potential of using an _E. coli_ strain with a severed TCA cycle for biotechnological applications. RESULTS GROWTH RECOVERY OF TCA CYCLE-DEFICIENT _E. COLI_ AFTER ALE Adaptive laboratory
evolution was performed on the TCA cycle-deficient _E. coli_ strain dTCA (BW25113 _ΔaceAΔsucAΔgadAΔgadBΔpoxB_::_acs_, Supplementary Table 1) to restore aerobic growth in glucose minimal
medium. The TCA cycle-deficient strain was pre-cultivated overnight in Luria-Bertani medium. For the first and second transfers, 5 mL culture was transferred into 50 mL glucose minimal
medium supplemented with 5 mL and 0.5 mL Luria-Bertani respectively. For the following serial passages, 0.5 mL culture was transferred into 50 mL glucose minimal medium without supplements.
The dTCA strain lost a key gene of TCA cycle, α-ketoglutarate dehydrogenase (_ΔsucA_), which encodes for the enzyme responsible for converting α-ketoglutarate to succinyl-CoA, as well as
genes for two bypass pathways, the glyoxylate shunt (_ΔaceA_) and γ-aminobutyric acid shunt (_ΔgadAΔgadB_). In order to monitor the acetate (acetyl-CoA) pool and determine if the TCA cycle
in the dTCA strain remained blocked during evolution, the pyruvate oxidase gene _poxB_ was replaced with the acetyl-CoA synthetase gene _acs_ (_ΔpoxB::acs_). In this situation, acetate
formed from acetyl-CoA was converted back to the acetyl-CoA pool. Adaptive laboratory evolution was conducted by continuous aerobic culture in glucose minimal medium. After 48 days of
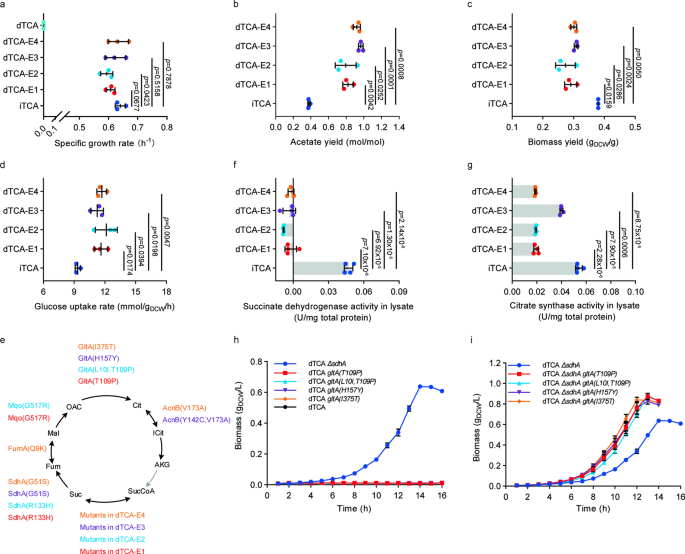
evolution (~230 generations), cells regained the ability to grow aerobically in glucose minimal medium. The specific growth rates of two evolutionary endpoint strains (µ = 0.61 ± 0.02 h−1
for dTCA-E1, µ=0.59 ± 0.02 h−1 for dTCA-E2) were similar to that of a control strain iTCA with an intact TCA cycle (BW25113 _ΔpoxB_::_acs_, µ=0.64 ± 0.02 h−1) (Fig. 1a). The acetate yields
of dTCA-E1 (0.82 ± 0.06 mol/mol) and dTCA-E2 (0.79 ± 0.12 mol/mol) during log-phase were significantly higher than that of iTCA strain (0.38 ± 0.01 mol/mol, Fig. 1b). When ALE was repeated,
similar results were obtained. Two evolutionary endpoint strains had comparable growth rates (0.62 ± 0.04 h−1 for dTCA-E3, 0.63 ± 0.04 h−1 for dTCA-E4) and increased acetate yields (0.97 ±
0.03 mol/mol for dTCA-E3, 0.92 ± 0.04 mol/mol for dTCA-E4) (Fig. 1a, b). The biomass yields of all evolutionary endpoint strains were significantly lower than that of the iTCA strain (Fig.
1c), while the glucose uptake rates of those four evolved strains were significantly higher than that of iTCA strain (Fig. 1d). MUTATIONS IN GENES ENCODING TCA CYCLE ENZYMES IN EVOLVED _E.
COLI_ Whole genome sequencing (https://www.ncbi.nlm.nih.gov/sra/PRJNA967749) and mutations analysis (the result of genomic mutations analysis was in Supplementary Data 1–6) were performed on
the evolutionary endpoint strains and the unevolved dTCA strain to uncover the mechanisms behind growth recovery. The analysis of gene mutations in the evolved strains revealed common
mutations in genes encoding enzymes in the TCA cycle (Fig. 1e, Supplementary Data 6). Specifically, genes encoding for succinate dehydrogenase subunit A (_sdhA_) and citrate synthase
(_gltA)_ were mutated in all four evolutionary endpoint strains. To investigate how these mutations affected enzyme activity, the evolved strains were cultivated in glucose medium to
log-phase and enzymatic activity of succinate dehydrogenase and citrate synthase were measured. No significant succinate dehydrogenase activity was detected in the lysate of the four evolved
strains containing succinate dehydrogenase mutations (Fig. 1f). The activities of GltA(T109P) (0.019 ± 0.002 U/mg total protein), GltA(L10I, T109P) (0.019 ± 0.001 U/mg total protein),
GltA(H157Y) (0.040 ± 0.001 U/mg total protein) and GltA(I375T) (0.019 ± 0.000 U/mg total protein) were significantly attenuated compared to the control strain iTCA (0.054 ± 0.003 U/mg total
protein) (Fig. 1g). These results suggested that inactivation of succinate dehydrogenase and attenuation of citrate synthase were critical for growth recovery of TCA cycle defected _E.
coli_. To further test this hypothesis, _sdhA_ was deleted in the unevolved TCA cycle-deficient starting strain dTCA and _gltA_ was replaced with the identified _gltA_ mutants (GltA(T109P),
GltA(L10I, T109P), GltA(H157Y) and GltA(I375T)). As shown in Fig. 1h, deletion of _sdhA_ was sufficient to restore aerobic growth (μ = 0.35 ± 0.01 h−1, Supplementary Table 2), which was
consistent with the previous work13. While mutations of _gltA_ alone could not restore aerobic growth of dTCA strain (Fig. 1h, Supplementary Table 2), they did significantly improve aerobic
growth rate of the strain dTCA _ΔsdhA_ (Fig. 1i, Supplementary Table 2). These results suggested that succinate dehydrogenase inactivation was essential for the growth recovery of TCA cycle
defected _E. coli_, while the attenuation of citrate synthase was beneficial but not necessary. METABOLISM REWIRING IN EVOLVED TCA CYCLE-DEFICIENT _E. COLI_ To elucidate the metabolic
mechanisms for growth recovery of evolved TCA cycle-deficient _E. coli_, 13C-metabolism flux analysis (13C-MFA) was performed on the evolved dTCA-E1 strain and iTCA strain, which had a
complete TCA cycle (Data of mass isotopomer distributions were listed in Supplementary Data 7, Data of MFA were shown in Supplementary Data 8). The 13C-MFA results uncovered extensive
metabolic flux rewiring all throughout central carbon metabolism (Fig. 2a), and provided additional support for the genome sequencing results and in vitro enzyme activity measurements. In
the evolved dTCA-E1 strain, no metabolic flux was detected from α-ketoglutarate to succinyl-CoA (0 ± 0), consistent with the knockout of _sucA_ gene; and from succinate to fumarate (0 ± 0),
consistent with the presumed inactivation of SdhA (R133H) in dTCA-E1 strain (Fig. 1f). Moreover, citrate synthase flux was significantly reduced in dTCA-E1 (7 ± 0) compared to iTCA strain
(27 ± 1), consistent with citrate synthase activity attenuation (Fig. 1g). In the dTCA-E1 strain, succinyl-CoA was regenerated exclusively from succinate, while in the iTCA strain all of
succinyl-CoA was derived from the decarboxylation of α-ketoglutarate in the TCA cycle. As SdhA was inactivated, succinate pool in dTCA-E1 might be supplemented from fumarate by L-aspartate
oxidase (NadB)14,15 or fumarate reductase (Frd)16 (Supplementary Fig. 1). The low regeneration flux of succinyl-CoA in dTCA-E1 (2 ± 0) compared to iTCA (14 ± 1) further suggested that this
step may be a metabolic bottleneck for cell growth given that succinyl-CoA is required for the biosynthesis of several amino acids in _E. coli_17. In the dTCA-E1 strain, only 2.2% of carbon
was converted into CO2 via the TCA cycle, whereas in the iTCA strain, this percentage was much higher at 11.7% (Fig. 2b, Supplementary Data 8). Notably, CO2 production in the dTCA-E1 strain
was primarily associated with the synthesis of α-ketoglutarate. Most of the CO2 released by dTCA-E1 was generated during the formation of acetate from pyruvate (Supplementary Data 8). In
addition to altered TCA cycle flux patterns, significant rewiring of upper metabolism was observed in the evolved strain. The oxidative pentose phosphate pathway (oxPPP) flux was
significantly upregulated in dTCA-E1 compared to iTCA (37 ± 0 vs. 24 ± 1) (Fig. 2a). As a result, more than 85% of NADPH required for cell growth was produced via oxPPP in dTCA-E1 compared
to less than 40% in iTCA (Fig. 2c, Supplementary Data 8). Remarkably, the transhydrogenase flux was negligible in dTCA-E1 strain (3 ± 3), whereas in iTCA the transhydrogenase flux was a
major contributor to biosynthetic NADPH production (Fig. 2c, Supplementary Data 8). The glycolytic flux to acetyl-CoA was significantly higher in dTCA-E1 than in iTCA (127 ± 1 vs. 105 ± 2).
This increased glycolysis flux, combined with reduced citrate synthase flux, produced a metabolic imbalance that was manifested as increased acetate secretion in dTCA-E1 compared to iTCA
(108 ± 1 vs. 60 ± 2). The remodeling of central carbon metabolism also affected energy generation. Compared to strain iTCA, dTCA-E1 obtained more ATP from substrate level phosphorylation and
acetate production (Fig. 2d, Supplementary Data 8) and showed higher overall ATP metabolism efficiency. The TCA cycle of dTCA-E1 neither directly generate ATP (Fig. 2d) nor produce
NADH/FADH2 (Fig. 2e) for coupling with oxidative phosphorylation to produce ATP. Since the absolute glucose uptake rate of dTCA-E1 strain (11.7 mmol/gDCW/h) was significantly higher than
that of iTCA (9.4 mmol/gDCW/h) (Fig. 2a, Supplementary Table 3), we determined that the absolute ATP production rate was nearly 40% higher in dTCA-E1 strain (113 mmol/gDCW/h) than in iTCA
strain (83 mmol/gDCW/h) (Supplementary Fig. 2c, Supplementary Data 8). These results suggested that the TCA cycle was not important for energy generation in dTCA-E1, in which ATP metabolism
was efficiently rebalanced through coordinated remodeling of central carbon metabolism. GROWTH RECOVERY BY ELIMINATING THE REQUIREMENT FOR SUCCINYL-COA Based to the results of the metabolic
mechanism and genetic basis for aerobic growth recovery, we hypothesized that limitations in the supply of succinyl-CoA could be the cause for the inability of TCA cycle-deficient _E. coli_
to grow in glucose minimal medium. Succinyl-CoA is involved in the biosynthesis of methionine, meso-2,6-Diaminopimelate (meso-DAP) and lysine, and participates in tetrapyrroles and heme
synthesis17. When the TCA cycle was blocked at the node of α-ketoglutarate dehydrogenase, succinyl-CoA could only be synthesized from succinate. We reasoned that availability of succinyl-CoA
would be negatively correlated with succinate dehydrogenase activity that consumes succinate and thus there would be a strong selective pressure for inactivation of succinate dehydrogenase
during adaptive evolution. Succinate dehydrogenase mutations were found in all 30 randomly isolated clones (Supplementary Tables 4–6) when we sequenced the succinate dehydrogenase operon in
dTCA or BW25113 _ΔaceAΔsucA_ clones that survived on glucose M9 minimal medium plate. This finding indicated that succinate dehydrogenase mutation was seemingly inevitable. However, _E.
coli_ with defective succinate dehydrogenase can’t be used to produce chemicals like G-7-ADCA, as it requires the TCA cycle to regenerate α-ketoglutarate from succinate. Hence, we explored
an alternative strategy to circumvent the succinyl-CoA requirement. As heme and tetrapyrroles can be synthesized from glumatyl-tRNA instead of succinyl-CoA in _E. coli_18, we hypothesized
that the causal bottleneck for growth of TCA cycle-deficient _E. coli_ was the biosynthesis of methionine, meso-DAP and lysine due to insufficient succinyl-CoA supply. To test this
hypothesis, strain BW25113 _ΔaceAΔsucA_ was grown in glucose minimal medium supplemented with either succinate (2 mmol/L) or an amino acid mixture (1 mmol/L each of
meso-2,6-diaminoheptanedioate, lysine and methionine). In both media, strain BW25113 _ΔaceAΔsucA_ grew well, which was faster than strain BW25113 _ΔaceAΔsucAΔsdhA_ in M9 minimal medium (Fig.
3a, Supplementary Table 7). Sequencing of the _sdhCDAB_ gene locus in BW25113 _ΔaceAΔsucA_ strain during aerobic growth with supplements revealed no mutations of succinate dehydrogenase.
Additionally, the intracellular succinyl-CoA level in glucose-grown BW25113 _ΔaceAΔsucA_ strain supplemented with amino acid mixture (1 mmol/L each of DAP, lysine and methionine) was
measured as 208.21 ± 37.82 nmol/gDCW, which was significantly lower than glucose-grown BW25113 (499.28 ± 162.47 nmol/gDCW) (Fig. 3d, the intracellular succinyl-CoA levels were in
Supplementary Data 9). This result provided further evidence for our hypothesis that insufficient availability of succinyl-CoA was the cause for growth arrest in TCA cycle-deficient _E.
coli_. Some bacteria have evolved alternative metabolic pathways for the biosynthesis of methionine, meso-DAP and lysine that do not require succinyl-CoA (Supplementary Fig. 3)19,20,21. For
example, in _Bacillus subtilis_, lysine and methionine are synthesized through acetyl-CoA dependent synthesis pathways. We hypothesized that by introducing acetyl-CoA dependent enzymes from
_B. subtilis_ into _E. coli_, the aerobic growth of TCA cycle-deficient _E. coli_ might be recovered in glucose minimum medium. To test this, the acetyl-CoA dependent pathways for
biosynthesis of methionine, meso-DAP and lysine from _B. subtilis_ were introduced into _E. coli_ BW25113 resulting in _E. coli_ DDPYM strain (BW25113 _ΔdapD_::_dapH(Bs)-dapL(Bs)-patA(Bs)
ΔmetA_::_yjcI(Bs)-metA(Bs)_) (Fig. 3b, Supplementary Table 1). Strain DDPYM had a similar growth rate (0.54 ± 0.05 h−1) as wild-type _E. coli_ BW25113 (0.54 ± 0.06 h−1) in glucose minimal
medium (Fig. 3c, Supplementary Table 7). Next, genes involved in TCA cycle and glyoxylate shunt (_sucABCD_ and _aceA)_ were knocked-out in DDPYM resulting in strains DDPYM _ΔaceAΔsucA_ and
DDPYM _ΔaceAΔsucABCD_. Known succinyl-CoA synthesis pathways in DDPYM _ΔaceAΔsucABCD_ were whole removed (_ΔsucABCD_). The specific growth rates of DDPYM _ΔaceAΔsucA_ and DDPYM
_ΔaceAΔsucABCD_ were 0.51 ± 0.05 h−1 and 0.52 ± 0.04 h−1, respectively (Fig. 3c, Supplementary Table 7), which were significantly higher than that of strain BW25113 _ΔaceAΔsucAΔsdhA_ (0.32 ±
0.04 h−1) and similar to DDPYM and BW25113 (Fig. 3c, Supplementary Table 7). The levels of succinyl-CoA in strains DDPYM and DDPYM _ΔaceAΔsucA_ were determined to be 1.75 ± 0.73 nmol/gDCW
and 1.04 ± 0.06 nmol/gDCW, respectively (Fig. 3d, Supplementary Data 9). While, the succinyl-CoA level in strain DDPYM _ΔaceAΔsucABCD_ (strain ZH40) was too low to be accurately measured in
biological replicate tests (Fig. 3d, Supplementary Data 9). These results confirm that growth arrest observed in TCA cycle-deficient _E. coli_ strains was indeed the results of limitations
in succinyl-CoA supply and not energy supply. In short, by introducing heterogeneous acetyl-CoA dependent pathways for biosynthesis of methionine, meso-DAP and lysine, _E. coli_ could
successfully grow with a severed TCA cycle (_ΔaceAΔsucA_ or _ΔaceAΔsucABCD_), which was enabled by eliminating the requirement for succinyl-CoA in amino acid biosynthesis pathways17,22. TCA
CYCLE-DEFICIENT _E. COLI_ AS AN EFFICIENT AEROBIC CHASSIS To demonstrate that TCA cycle-deficient DDPYM can serve as a versatile and efficient chassis strain for aerobic fermentations
without requirement of succinyl-CoA for amino acid (methionine, meso-DAP and lysine) biosynthesis, strain ZH40 (DDPYM _ΔaceAΔsucABCD_) without succinyl-CoA synthesis pathways (_sucAB_ and
_sucCD_)22 was constructed and engineered for biosynthesis of several chemicals (Fig. 4a). In previous work, we used a TCA cycle-deficient _E. coli_ (_ΔaceAΔsucA_) to convert penicillin G to
G-7-ADCA by overexpressing deacetoxycephalosporin C synthase (DAOCS)12. As penicillin G and G-7-ADCA was toxic to _E. coli_, the defective TCA cycle could not be repaired with
α-ketoglutarate-dependent DAOCS to support cell growth in glucose minimal medium. Here, DAOCS was overexpressed in strain ZH40, which can grow aerobically on glucose minimal medium. Protein
expression of DAOCS in ZH40 was similar to that in BW25113 and DDPYM background strains with an intact TCA cycle (Fig. 4b). Notably, G-7-ADCA production in ZH40 host strain (7.02 ± 0.27
mmol/L) was more than 3-fold higher compared to BW25113 (2.20 ± 0.31 mmol/L) and DDPYM background strains (1.45 ± 0.05 mmol/L) (Fig. 4c). Next, strain ZH40 was engineered to produce
glutamate, one of the twelve top value-added chemicals from biomass. By overexpressing phosphoenolpyruvate carboxylase (_ppc_) and glutamate dehydrogenase (_gdhA_) (Fig. 4a), 5 mmol/L
glutamate was obtained in 17 h during aerobic growth on 20 mmol/L glucose (Fig. 4d, Supplementary Fig. 4a) by strain ZH40-gdhA-ppc, while no detectable glutamate was observed in strains with
an intact TCA cycle BW25113-gdhA-ppc and DDPYM-gdhA-ppc strains (Fig. 4d, Supplementary Fig. 4a). In the whole-cell biocatalysis, little amount of AKG were detected in BW25113 and DDPYM
(Supplementary Fig. 5a, b), while strain ZH40 (DDPYM _ΔaceAΔsucABCD_) with defective TCA cycle produced 14.93 ± 0.50 mmol/L AKG after complete consumption of 50 mmol/L glucose (Supplementary
Fig. 5c). These results demonstrated the advantage of the defective TCA cycle on AKG accumulation and the TCA-cycle defective strain held potential in production of chemicals derived from
AKG. To demonstrate that strain ZH40 can also be used for efficient production of acetyl-CoA derived products, pyruvate oxidase gene (_poxB_) was knocked-out from strain ZH40, resulting in
strain ZH44 (DDPYM _ΔaceAΔsucABCDΔpoxB_) which could only generate acetate from acetyl-CoA. Strains ZH40 and ZH44 produced acetate at a higher yield and rate than _E. coli_ strains with an
intact TCA cycle (Fig. 4e, Supplementary Table 8), while maintaining a similar growth rate (0.63 ± 0.01 h–1 and 0.64 ± 0.00 h–1, respectively) as BW25113 (0.66 ± 0.01 h–1) and DDPYM (0.63 ±
0.00 h–1) (Supplementary Fig. 4b, Supplementary Table 8). When citrate synthase of ZH44 was mutated as GltA(T109P) to reduce flux of acetyl-CoA into TCA cycle, the growth rate, biomass
yield, acetate yield and acetate production rate of the resulting strain ZH45 (ZH44 _gltA(T109P)_) were increased during exponential growth phase (Fig. 4e, Supplementary Fig. 4b,
Supplementary Table 8). During whole-cell biocatalysis, acetate yield increased further to 0.96 ± 0.01 mol/mol in ZH40 and ZH44 (Supplementary Fig. 5c, d, Figs. 4f), and 1.42 ± 0.04 mol/mol
in ZH45 (Supplementary Fig. 5e, Fig. 4f), while no acetate was accumulated in cultures of _E. coli_ strains with an intact TCA cycle (Supplementary Fig. 5a, b, Fig. 4f). These results
demonstrated the benefit of the defective TCA cycle for production of chemicals from acetyl-CoA. In whole-cell catalysis stage, the resting cells did not grow. The yields of metabolites from
glucose in whole-cell catalysis stage (Supplementary Fig. 5) were used to analyze the distribution of metabolic flow (Fig. 4f). To calculate the fraction of carbon fluxes from glucose to
metabolites (Fig. 4f), the measured yields of metabolites were divided by the theoretical yields of their respective pathways, with CO2 and any unmeasured metabolites considered as lost
carbon. In the strains with complete TCA cycle, BW25113 and DDPYM, the TCA cycle decarboxylation caused complete carbon loss from glucose (Fig. 4f, Supplementary Fig. 5a, b). However, the
TCA cycle-deficient strains ZH40, ZH44, and ZH45 showed a significant reduction in carbon loss, with over 70% of the carbon flux diverted to acetate and α-ketoglutarate (AKG) (Fig. 4f,
Supplementary Fig. 5c–e). Mutation of citrate synthase in strain ZH45 leads to a decrease in AKG flux and an increase in acetate flux compared to strain ZH44 (Fig. 4f, Supplementary Fig. 5d,
e). However, these TCA cycle defective strains ZH40, ZH44, and ZH45 also presented above 20% carbon loss. Citrate synthase mutation caused a modest increase in lost carbon in strain ZH45
(26.54 ± 2.17%) comparing to strain ZH44 (20.44 ± 0.92%) (Fig. 4f). We hypothesized that there might exist carbon loss from acetyl-CoA or pyruvate in strain ZH45, which might be decreased if
carbon flux was directed from pyruvate to pyruvate-derived chemical such as acetoin. Acetoin can be synthesized from pyruvate using acetolactate synthase (AlsS) and acetolactate
decarboxylase (AlsD) without production or consumption of reductants or energy (Fig. 4a). Genes _alsS_ and _alsD_ from _Bacillus subtilis_ were introduced into strain ZH42, resulting in
strain ZH42-alsSD (Supplementary Table 1). In whole-cell catalysis, strain ZH42-alsSD generated 35.98 ± 0.57 mmol/L acetoin from 50 mmol/L glucose, which was 72% of the theoretical maximal
yield from glucose (Supplementary Fig. 5f). Only 0.17 ± 0.67% of the carbon in ZH42-alsSD was lost from glucose, and most of carbon flowed to metabolites including pyruvate (4.14 ± 0.94%),
acetoin (71.95 ± 1.13%), acetate (11.65 ± 0.87%), and α-ketoglutarate (12.09 ± 0.35%) (Fig. 4f). These results demonstrated the significant potential of TCA cycle-deficient DDPYM strains for
minimizing carbon dissipation and facilitating chemicals production. DISCUSSION The conventional TCA cycle in heterotrophic microbes has been frequently modified to improve chemicals
production in aerobic fermentation1,2,3,4,12,23,24. While some microorganisms such as cyanobacteria25, methylotrophic bacteria26,27 and _Mycobacterium tuberculosis_28,29 have been found to
have an incomplete TCA cycle. However, it was found that a break in the TCA cycle seriously affects aerobic growth of _E. coli_ in glucose minimal medium1,2,3. In this work, we successfully
reprogramed metabolism of _E. coli_ without a complete TCA cycle to grow well on glucose and convert glucose into products at high yields. Adaptive laboratory evolution was used to gain a
better understanding of the underlying mechanisms causing the growth defect observed when TCA cycle was severed at the α-ketoglutarate dehydrogenase step in _E. coli_. It was discovered that
the limiting factor for growth was not energy limitation but insufficient supply of succinyl-CoA resulting in the inability to produce three amino acids, methionine, meso-DAP and lysine.
Succinyl-CoA can only be generated from succinate in _E. coli_ if the α-ketoglutarate dehydrogenase reaction was blocked (_ΔsucA_). When the glyoxylate cycle was also blocked by inactivation
of isocitrate lyase (_ΔaceA_), the _E. coli_ with defective TCA-cycle (_ΔaceAΔsucA_) lacked sufficient succinate to maintain the succinyl-CoA pool. Although succinate might be supplemented
from fumarate by L-aspartate oxidase (NadB)14,15 or fumarate reductase (Frd)16 (Supplementary Fig. 1), the amount of succinate was limited. Due to succinate being rapidly oxidized by
succinate dehydrogenase, TCA cycle defected (_ΔaceAΔsucA_) _E. coli_ could not maintain a sufficient succinyl-CoA pool without enough intracellular succinate, unless its succinate
dehydrogenase was mutated. Upon transfer to glucose minimal medium plates from rich medium, those survived dTCA and BW25113 _ΔaceAΔsucA_ clones were found to have succinate dehydrogenase
mutations (Supplementary Tables 4–6). This work provides evidence that succinate dehydrogenase mutation is necessary and unavoidable for recovering aerobic growth of TCA-cycle defective _E.
coli_ (_ΔaceAΔsucA_) in glucose minimal medium. However, succinate dehydrogenase mutation was inapplicable to the production of chemicals such as G-7-ADCA, which required a reconstituted TCA
cycle to regenerate α-ketoglutarate from succinate12. Therefore, we attempted to circumvent succinyl-CoA and construct a more versatile chassis. It is a challenge to bypass succinyl-CoA
requirement, for succinyl-CoA has always been considered as one of the twelve essential precursor metabolites for cell growth17. This work showed that growth could be recovered by bypassing
the requirement for succinyl-CoA in amino acids synthesis through expression of heterologous acetyl-CoA dependent enzymes. Succinyl-CoA synthesis pathways genes _sucAB_ and _sucCD_ were
knocked out in TCA cycle-deficient _E. coli_ strain ZH40, which still retained the ability to provide essential precursor metabolites and energy for aerobic growth (Fig. 3c, Supplementary
Fig. 4b) and over expression of protein (Fig. 4b) while avoiding unnecessary oxidization of acetyl-CoA to CO2 in the TCA cycle (Fig. 4e, f, Supplementary Fig. 5c). The succinyl-CoA level in
ZH40 was hundreds of times lower than that of wild type _E. coli_ BW25113, making it too low to determine accurately (Fig. 3d, Supplementary Data 9). Citrate synthase catalyzes the critical
irreversible step that controls the amount of the acetyl-CoA pool and the metabolic flux into TCA cycle. In this study, mutations in citrate synthase reducing the enzyme activity of citrate
synthase were found beneficial for the growth recovery of TCA cycle-deficient (_ΔaceAΔsucA_) _E. coli_ (Fig. 1i, Supplementary Table 2, Supplementary Table 8). Tricarboxylic acid metabolic
flux was decreased in evolved strain dTCA-E1, comparing to unevolved iTCA strain (Fig. 2a). The mutation in the citrate synthase of strain dTCA-E1 was confirmed able to reduce
α-ketoglutarate secretion of TCA cycle-deficient (_ΔaceAΔsucA_) _E. coli_ (Fig. 4f, Supplementary Fig. 5e). A previous work reported that after evolution of the _sdhCB_ knock-out _E. coli_,
mutations in α-ketoglutarate dehydrogenase were found that reduced tricarboxylic acid metabolic flux and succinate secretion30. According to the founding of previous reported work and this
study, it was assumed that a glucose-grown TCA cycle-defective _E. coli_ strain would tend to minimize carbon waste by reducing the citrate (tricarboxylic acid) metabolism during evolution.
For a heterotrophic microbe like _E. coli_, glucose is entirely oxidized to CO2 and H2O through the TCA cycle and respiratory chain in aerobic conditions. However, complete oxidation of each
acetyl-CoA via the TCA cycle leads to the emission of two CO2, which reduces the carbon flux directed to product formation and negatively affects product yield. Therefore, eliminating the
TCA cycle in _E. coli_ can decrease carbon dissipation and improve the efficiency of chemicals biosynthesis. To prove this concept, the TCA cycle-deficient DDPYM strain was developed and
successfully used to achieve higher yield biosynthesis of chemicals from acetyl-CoA (acetate) and AKG point (G-7-ADCA, glutamate) when compared to strains with an intact TCA cycle (Fig. 4,
Supplementary Fig. 5). On the other hand, _E. coli_ strains with an intact TCA cycle couldn’t prevent carbon loss from acetyl-CoA oxidation, even when their growth is strictly restricted in
a whole-cell catalysis solution (Fig. 4e, f, Supplementary Fig. 5a, b). The chassis strain ZH40, with the genotype DDPYM _ΔaceAΔsucABCD_, accumulated α-ketoglutarate (Fig. 4f, Supplementary
Fig. 5c), making it suitable for the efficient biosynthesis of various products that require α-ketoglutarate, such as G-7-ADCA, hydroxy-proline, hydroxy-isoleucine, and glutamate family
amino acids. On the other hand, the chassis strain ZH42, with the genotype DDPYM _ΔaceAΔsucABCDΔpoxBΔpta gltA(T109P)_, is a versatile chassis for producing chemicals derived from acetyl-CoA
(e.g., butanol and fatty acids) as well as chemicals derived from glycolysis pathway intermediates (e.g., pyruvate). In summary, this work opens up an exciting avenues for more efficient
biosynthesis of industrial chemicals in the future. METHODS PLASMID AND STRAIN CONSTRUCTION Plasmids information and strains genotypes were listed in Supplementary Table 1. Gene knock-in and
knock-out experiments were conducted with reported system31,32,33,34,35,36,37. Strain dTCA in this work was constructed via deleting genes _gadA_ and _gadB_ from strain PG0512. The DAOCS
gene was cloned from plasmid pDB1S-H712 and inserted into plasmid pET28b(+) between _Nco_I and _Xho_I sites to form plasmid pET28b(+)-DAOCS (Supplementary Table 1). For overexpression of
DAOCS, a section of DE3 cassette (DE3’) expressing T7 RNA polymerase was cloned from _E. coli_ BL21(DE3) and integrated into the chromosome of the host strain. Genes _gdhA_ and _ppc_ were
cloned from _E. coli_ BW25113 genome and inserted into plasmid pSC2s under P_tac_ promoter to form plasmid pSC2s-gdhA-ppc (Supplementary Table 1). Genes _alsS_ and _alsD_ were cloned from
_Bacillus subtilis_ 168 genome to construct a synthetic operon P_tac_-_alsSD_ and insert into the _ldhA_ locus in strain ZH42-alsSD by the CRISPR system32. Details for genome and plasmids
engineering were listed in Supplementary Data 10. ADAPTIVE LABORATORY EVOLUTION Adaptive laboratory evolution was performed via a series of passages in 50 mL glucose minimal medium (50 mL of
M9 minimal medium with 20 mmol/L glucose) in 100 mL flasks shaking at 200 rpm and 37 °C. The TCA cycle-deficient strains pre-cultured overnight in Luria-Bertani medium were transferred to
glucose minimal medium. For the first and second transfers, 5 mL culture was transferred into 50 mL minimal medium supplemented with 5 mL and 0.5 mL Luria-Bertani respectively. For the
following serial passages, 0.5 mL culture was transferred into 50 mL glucose minimal medium without supplements. Endpoint strains were isolated from the evolutionary population via streaking
on glucose minimal medium plate when the growth rate did not increase any more. Clones isolated from the plate were frozen in 150 g/L glycerol and stored at –80 °C for future analysis, such
as growth analysis, enzyme activities and genome sequencing. During ALE experiments, GREACE was used to accelerate mutation rate38, and the relevant mutation plasmid was eliminated during
adaptive laboratory evolution without antibiotic selective pressure. MUTATIONS ANALYSIS The genome DNA of the evolved strains was extracted and prepared as Illumina PE150 libraries. It was
then sequenced using Illumina HiSeq X technology by MajorBio company. The BWA-Samtools-Bcftools SNP calling pipeline39,40 was employed to identify mutations, with the _E. coli_ BW25113
genome (Accession: CP009273.1) serving as the reference sequence. Only variations that met the criteria of being covered by more than 50 reads and located in the ORF or the upstream 100 bp
untranslated region were retained. The genome of unevolved dTCA was also sequenced and analyzed to eliminate any innate genetic variations before ALE experiment. The reads associated with
this study have been deposited in the National Center for Biotechnology Information (NCBI) databank SRA, with the BioProject accession number (Accession: PRJNA967749). SNP analysis for the
four evolutionary endpoint strains and unevolved dTCA were listed in Supplementary Data 1–6. Survived clones from two groups of ALE experiments (ALE-1 and ALE-2) were randomly isolated by
streaking on glucose M9 minimal medium plates after the second transfer of evolutionary populations in glucose M9 minimal medium supplemented with 0.5 mL Luria-Bertani. For mutation analysis
of citrate synthase and succinate dehydrogenase, clones that survived on glucose M9 minimal medium plates were randomly selected for PCR reactions and Sanger sequencing. BW25113
_ΔaceAΔsucA_, which was pre-cultivated in Luria-Bertani medium, was washed twice with M9 minimal medium and spread on glucose M9 minimal medium plates to isolate surviving clones. The
results of the mutation analysis for citrate synthase and succinate dehydrogenase are listed in Supplementary Tables 4–6. GROWTH AND METABOLITE ANALYSIS Unless otherwise noted, growth assays
were conducted with 50 mL cultures in 250 mL flasks shaking at 200 rpm and 37 °C. All strains grew in M9 minimal medium with 20 mmol/L glucose and were monitored by measuring the optical
density at 600 nm (OD600) using a spectrophotometer (Eppendorf BioPhotometer). The OD600 values were converted to cell dry weight concentrations using a determined OD600-dry cell weight
relationship for _E. coli_ (1.0 OD600 = 0.32 gDCW/L41). The initial OD600 of the inoculated cultures was 0.01. If a strain was pre-cultivated in Luria-Bertani medium, it should be washed
twice with M9 minimal medium before inoculation. Supplements was added as indicated in the text if it’s used. Growth rate was calculated using linear regression of the natural logarithm of
OD600 versus time. Biomass (or metabolites) yield was calculated using linear regression of biomass dry weight (or metabolites concentration) and glucose concentration in the medium. Glucose
uptake rate (mmol/gDCW/h) was determined as the ratio of growth rate and biomass yield. The production per gram of dry cell weight (mmol/gDCW) was determined using linear regression on
metabolite concentrations and biomass dry weight during log phase of growth. After centrifugation of the samples, the concentrations of glucose, pyruvate, acetoin, acetate and
α-ketoglutarate in the supernatants were determined by high-performance liquid chromatography (HPLC, Agilent 1260 series, Hewlett–Packard), equipped with a 300 mm × 7.8 mm Aminex HPX-87H
column (Bio-Rad, Hercules, CA, USA) and a refractive index detector. Analysis was performed at 55 °C with a mobile phase of 5 mmol/L H2SO4 at a flow rate of 0.6 mL/min. Concentrations of
glutamate, G-7-ADCA and penicillin G were measured by HPLC (Agilent 1260 series, Hewlett–Packard), equipped with a ZORBAX Eclipse XDB-C18 column (4.6 mm × 150 mm, 5 μm; Agilen) and a
diode-array detector. Glutamate was monitored at 360 nm after DNFB derivation42, G-7-ADCA and penicillin G were respectively measured at 220 nm and 260 nm12. DETERMINATION OF INTRACELLULAR
SUCCINYL-COA LEVEL Strains were cultivated in 20 mmol/L glucose M9 minimal medium to measure the intracellular succinyl-CoA level. Then, at the log phase of growth, the strains were
collected by centrifugation at 12,000 × _g_ for 5 min. Subsequently, the collected strains were suspended (OD600 = 20) in 1 mL of 80% (vol/vol) methanol (–80 °C) for complete ultrasonication
at 4 °C. The resulting supernatant was collected and dried without heat in SpeedVac, while the pellet was redissolved in deionized water. The succinyl-CoA concentration was determined in
the redissolved pellet using LC-MS/MS method35. WHOLE-CELL CATALYSIS Whole-cell catalysis for glucose (50 mmol/L) catabolism was performed in 100 mL flasks at 37 °C and 200 rpm. The cells in
the log-phase were collected by centrifugation at 5000 × _g_ for 5 min at 4 °C and suspended in 10 mL Tris-MOPS buffer (100 mmol/L Tris-base and 100 mmol/L MOPS, pH 7.4) at a concentration
of about 3.2 gDCW/L. The whole-cell catalysis for producing G-7-aminodeacetoxycephalosporanic acid (G-7-ADCA) from penicillin G was conducted with glucose12. After 10 h of whole-cell
catalysis, the concentrations of acetate, acetoin, pyruvate, and α-ketoglutarate were measured to calculate their molar yields from glucose. The measured molar yields were then divided by
their pathway theoretical molar yields and utilized to track carbon flux from glucose to metabolites (fractions of carbon fluxes from glucose to metabolites were calculated and listed in the
source data of Fig. 4f). MOPS in the reaction buffer was used as an internal reference for the precise determination of metabolite concentrations, especially when the volume of the
whole-cell catalysis was changed. ENZYME ACTIVITY ANALYSIS For enzyme activity analysis, cells in log-phase were collected by centrifugation at 5000 × _g_ for 5 min at 4°C, suspended in 100
mmol/L Tris-Cl (pH 7.4) buffer, and lysed by sonication. For succinate dehydrogenase assays, 200 μL reaction solution contained 100 mmol/L Tris-Cl (pH 7.4), 2 mmol/L MgCl2, 4 mmol/L sodium
succinate, 2 mmol/L dichlorophenolindophenol, and 10 μL of cell lysate. The linear regression coefficient of OD600 value versus time (_R__2_ > 0.99) was used to determine succinate
dehydrogenase activity. For citrate synthase assays, 200 μL reaction solution contained 100 mmol/L Tris-Cl (pH 7.4), 2 mmol/L MgCl2, 2 mmol/L oxaloacetic acid, 1 mmol/L acetyl-CoA, 0.65
mmol/L 5,5’-dithiobis-(2-nitrobenzoic acid), and 10 μL of cell lysate. The linear regression coefficient of OD412 value versus time (_R__2_ > 0.99) was used to determine citrate synthase
activity. The enzyme activity unit (U) was defined as the amount of enzyme required to catalyze the conversion of 1 μmol substrate at room temperature within 1 min. Protein quantification
was conducted with coomassie brilliant blue protein assay, for which BSA was used as standard protein sample. CHEMICALS AND REAGENTS All chemicals and media for 13C-metabolic flux analysis
were purchased from Sigma-Aldrich (St. Louis, MO). Isotopic tracers were purchased from Cambridge Isotope Laboratories (Tewksbury, MA): [1,2-13C]glucose (99.5%), [1,6-13C]glucose (99.5%
13C), and [4,5,6-13C]glucose (99.5% 13C). The isotopic purity and enrichment of tracers were validated by GC-MS analysis as described43. Wolfe’s minerals (Cat. No. MD-TMS) and Wolfe’s
vitamins (Cat. No. MD-VS) were purchased from ATCC (Manassas, VA). The M9 minimal medium used for strains aerobic growth was supplemented with 10 mL/L of Wolfe’s minerals and 4.5 mg/L of
vitamin B1. Glucose was added as indicated in the text. All media and stock solutions were sterilized by filtration. Commercial reagents for other experiments were purchased from Shanghai
Yuanye Bio-Technology Co., Ltd. TRACER EXPERIMENTS For tracer experiments, cells were first precultured overnight at 37 °C in shaker flasks. Cells were then resuspended in fresh medium
containing a particular glucose tracer, either [1,2-13C]glucose, [1,6-13C]glucose, or [4,5,6-13C]glucose (10 mmol/L initial concentration). Three parallel labeling experiments were conducted
for each strain. The initial OD600 of the inoculated cultures was 0.015. After inoculation, cells were grown at 37 °C in parallel mini-bioreactors with a working volume of 10 mL44. Air was
sparged into the liquid at a rate of 12 mL/min to provide oxygen and to ensure sufficient mixing of the culture by the rising gas bubbles. Cells were collected for GC-MS analysis during the
mid-exponential growth phase when the biomass concentration (OD600) was between 0.5 and 0.7. GAS CHROMATOGRAPHY-MASS SPECTROMETRY GC-MS analysis was performed on an Agilent 7890B GC system
equipped with a DB-5MS capillary column (30 m, 0.25 mm i.d., 0.25 µm-phase thickness; Agilent J&W Scientific), connected to an Agilent 5977 A Mass Spectrometer operating under ionization
by electron impact (EI) at 70 eV. Helium flow was maintained at 1 mL/min. The source temperature was maintained at 230 °C, the MS quad temperature at 150 °C, the interface temperature at
280 °C, and the inlet temperature at 250 °C. Proteinogenic amino acids was analyzed by GC-MS after _tert_-butyldimethylsilyl (TBDMS) derivation45. Labeling of glucose (derived from glycogen)
and ribose (derived from RNA) were determined by GC-MS46,47. In all cases, mass isotopomer distributions were obtained by integration and corrected for natural isotope abundances48.
Measurement errors of 0.3% were assumed for all measured mass isotopomers49. METABOLIC NETWORK MODEL AND 13C-METABOLIC FLUX ANALYSIS The metabolic network model used for 13C-MFA is provided
in Supplementary Note 1. The model is based on the _E. coli_ model50, which includes all major metabolic pathways of central carbon metabolism, lumped amino acid biosynthesis reactions, and
a lumped biomass formation reaction. Updates to the model include making the reaction between PEP and pyruvate reversible51, and modeling the dilution of isotopic labeling by atmospheric
CO252. 13C-MFA calculations were performed using Metran software53, which is based on the elementary metabolite units (EMU) framework54. Fluxes were estimated by minimizing the
variance-weighted sum of squared residuals (SSR) between the measured and model predicted mass isotopomer distributions and acetate yield using non-linear least-squares regression. All
measured mass isotopomers are provided in Supplementary Data 7. For integrated analysis of parallel labeling experiments, the data sets were fitted simultaneously to a single flux
model52,55. Flux estimation was repeated 10 times starting with random initial values for all fluxes to find a global solution. At convergence, accurate 95% confidence intervals were
computed for all estimated fluxes by evaluating the sensitivity of the minimized SSR to flux variations. Precision of estimated fluxes was determined as follows56:
$${{{{{\rm{Flux}}}}}}\,{{{{{\rm{precision}}}}}}({{{{{\rm{stdev}}}}}})=[({{{{{{\rm{flux}}}}}}}_{{{{{{\rm{upper}}}}}} \, {{{{{\rm{bound}}}}}} \, 95\%}) -
({{{{{{\rm{flux}}}}}}}_{{{{{{\rm{lower}}}}}} \, {{{{{\rm{bound}}}}}} \, 95\%})]/4$$ (1) To describe fractional labeling of metabolites, G-value parameters were included in 13C-MFA. The
G-value represents the fraction of a metabolite pool that is produced during the labeling experiment, while 1-G represents the fraction that is naturally labeled, i.e. from the inoculum57.
By default, one G-value parameter was included for each measured metabolite in each data set. Reversible reactions were modeled as separate forward and backward fluxes. Net and exchange
fluxes were determined as follows: $${{{{{{\rm{v}}}}}}}_{{{{{{\rm{net}}}}}}}={{{{{{\rm{v}}}}}}}_{{{{{{\rm{f}}}}}}}-{{{{{{\rm{v}}}}}}}_{{{{{{\rm{b}}}}}}}$$ (2)
$${{{{{{\rm{v}}}}}}}_{{{{{{\rm{exch}}}}}}}=\min ({{{{{{\rm{v}}}}}}}_{{{{{{\rm{f}}}}}}},{{{{{{\rm{v}}}}}}}_{{{{{{\rm{b}}}}}}})$$ (3) To determine the goodness-of-fit, 13C-MFA fitting results
were subjected to a χ2-statistical test. In short, assuming that the model is correct and data are without gross measurement errors, the minimized SSR is a stochastic variable with a
χ2-distribution56. The number of degrees of freedom is equal to the number of fitted measurements _n_ minus the number of estimated independent parameters _p_. The acceptable range of SSR
values is between χ2α/2(_n_-_p_) and χ21-α/2(_n_-_p_), where α is a certain chosen threshold value, for example 0.05 for the 95% confidence interval. The transhydrogenase flux and oxidative
phosphorylation fluxes in the model are determined using the carbon fluxes estimated from 13C-MFA58. In short, in the metabolic model used for 13C-MFA, all reactions are redox balanced. The
assignment whether NADH, FADH2 or NADPH is oxidized or reduced in any reaction is based on the known biochemistry of _E. coli_ enzymes. The total production and consumption fluxes for each
cofactor are calculated from 13C-MFA results and then balanced. To balance NADPH fluxes, a transhydrogenase reaction is included in the metabolic model. To balance NADH and FADH2 fluxes,
oxidative phosphorylation reactions are included in the model. The results of flux analysis and cofactor balance analysis were listed in Supplementary Data 8. REPORTING SUMMARY Further
information on research design is available in the Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY Reads of genome sequencing have been deposited in the National
Center for Biotechnology Information (NCBI) databank SRA with the BioProject accession PRJNA967749. Source data are provided with this paper. REFERENCES * Smirnov, S. V. et al. Metabolic
engineering of _Escherichia coli_ to produce (2S, 3R, 4S)-4-hydroxyisoleucine. _Appl. Microbiol. Biotechnol._ 88, 719–726 (2010). Article CAS PubMed Google Scholar * Theodosiou, E. et
al. An artificial TCA cycle selects for efficient α-ketoglutarate dependent hydroxylase catalysis in engineered _Escherichia coli_. _Biotechnol. Bioeng._ 114, 1511–1520 (2017). Article CAS
PubMed Google Scholar * Zhang, H. L. et al. Efficient production of trans-4-Hydroxy-l-proline from glucose by metabolic engineering of recombinant _Escherichia coli_. _Lett. Appl.
Microbiol._ 66, 400–408 (2018). Article CAS PubMed Google Scholar * Soma, Y., Fujiwara, Y., Nakagawa, T., Tsuruno, K. & Hanai, T. Reconstruction of a metabolic regulatory network in
_Escherichia coli_ for purposeful switching from cell growth mode to production mode in direct GABA fermentation from glucose. _Metab. Eng._ 43, 54–63 (2017). Article CAS PubMed Google
Scholar * Portnoy, V. A., Bezdan, D. & Zengler, K. Adaptive laboratory evolution-harnessing the power of biology for metabolic engineering. _Curr. Opin. Chem. Biol._ 22, 590–594 (2011).
CAS Google Scholar * Dragosits, M. & Mattanovich, D. Adaptive laboratory evolution - principles and applications for biotechnology. _Micro. Cell Fact._ 12, 64 (2013). Article Google
Scholar * Winkler, J., Reyes, L. H. & Kao, K. C. Adaptive laboratory evolution for strain engineering. _Methods Mol. Biol._ 985, 211–222 (2013). Article CAS PubMed Google Scholar *
Aguilar, C. et al. Genetic changes during a laboratory adaptive evolution process that allowed fast growth in glucose to an _Escherichia coli_ strain lacking the major glucose transport
system. _BMC Genomics_ 13, 385 (2012). Article CAS PubMed PubMed Central Google Scholar * Chou, H. H., Marx, C. J. & Sauer, U. Transhydrogenase promotes the robustness and
evolvability of _E. coli_ deficient in NADPH production. _PLoS Genet._ 11, e1005007 (2015). Article PubMed PubMed Central Google Scholar * Charusanti, P. et al. Genetic basis of growth
adaptation of _Escherichia coli_ after deletion of _pgi_, a major metabolic gene. _PLoS Genet._ 6, e1001186 (2010). Article PubMed PubMed Central Google Scholar * Long, C. P., Gonzalez,
J. E., Feist, A. M., Palsson, B. O. & Antoniewicz, M. R. Dissecting the genetic and metabolic mechanisms of adaptation to the knockout of a major metabolic enzyme in _Escherichia coli_.
_Proc. Natl .Acad. Sci. USA_ 115, 222–227 (2018). Article CAS PubMed ADS Google Scholar * Lin, B. et al. Reconstitution of TCA cycle with DAOCS to engineer _Escherichia coli_ into an
efficient whole cell catalyst of penicillin G. _Proc. Natl. Acad. Sci. Usa._ 112, 9855–9859 (2015). Article CAS PubMed PubMed Central ADS Google Scholar * Creaghan, I. T. & Guest,
J. R. Succinate dehydrogenase-dependent nutritional requirement for succinate in mutants of _Escherichia coli_ K12. _J. Gen. Microbiol._ 107, 1–13 (1978). Article CAS PubMed Google
Scholar * Korshunov, S. & Imlay, J. A. Two sources of endogenous hydrogen peroxide in _Escherichia coli_. _Mol. Microbiol._ 75, 1389–1401 (2010). Article CAS PubMed PubMed Central
Google Scholar * Hermes, F. A. & Cronan, J. E. An NAD synthetic reaction bypasses the lipoate requirement for aerobic growth of _Escherichia coli_ strains blocked in succinate
catabolism. _Mol. Microbiol._ 94, 1134–1145 (2014). Article CAS Google Scholar * Hirsch, C. A., Davis, B. D., Rasminsky, M. & Lin, E. C. C. A fumarate reductase in _Escherichia coli_
distinct from succinate dehydrogenase. _J. Biol. Chem._ 238, 3770–3774 (1963). Article CAS PubMed Google Scholar * Noor, E., Eden, E., Milo, R. & Alon, U. Central carbon metabolism
as a minimal biochemical walk between precursors for biomass and energy. _Mol. Cell._ 39, 809–820 (2010). Article CAS PubMed Google Scholar * Jahn, D., Verkamp, E. & Soll, D.
Glutamyl-transfer RNA: a precursor of heme and chlorophyll biosynthesis. _Trends Biochem. Sci._ 17, 215–218 (1992). Article CAS PubMed Google Scholar * Scapin, G. & Blanchard, J. S.
Enzymology of bacterial lysine biosynthesis. _Adv. Enzymol._ 72, 279–324 (1998). CAS PubMed Google Scholar * Born, T. L. & Blanchard, J. S. Structure/function studies on enzymes in
the diaminopimelate pathway of bacterial cell wall biosynthesis. _Curr. Opin. Chem. Biol._ 3, 607–613 (1999). Article CAS PubMed Google Scholar * Zubieta, C., Arkus, K. A. J., Cahoon, R.
E. & Jez, J. M. A single amino acid change is responsible for evolution of acyltransferase specificity in bacterial methionine biosynthesis. _J. Biol. Chem._ 283, 7561–7567 (2008).
Article CAS PubMed Google Scholar * Yu, B. J. et al. _sucAB_ and _sucCD_ are mutually essential genes in Escherichia coli. _FEMS Microbiol Lett._ 256, 178–178 (2006). Article CAS
Google Scholar * Tovilla-Coutino, D. B., Momany, C. & Eiteman, M. A. Engineered citrate synthase alters acetate accumulation in _Escherichia coli_. _Metab. Eng._ 61, 171–180 (2020).
Article CAS PubMed Google Scholar * Harder, B. J., Bettenbrock, K. & Klamt, S. Temperature-dependent dynamic control of the TCA cycle increases volumetric productivity of itaconic
acid production by _Escherichia coli_. _Biotechnol. Bioeng._ 115, 156–164 (2018). Article CAS PubMed Google Scholar * Zhang, S. Y. & Bryant, D. A. The tricarboxylic acid cycle in
cyanobacteria. _Science_ 334, 1551–1553 (2011). Article CAS PubMed ADS Google Scholar * Wood, A. P., Aurikko, J. P. & Kelly, D. P. A challenge for 21st century molecular biology and
biochemistry: what are the causes of obligate autotrophy and methanotrophy? _FEMS Microbiol Rev._ 28, 335–352 (2004). Article CAS PubMed Google Scholar * Chistoserdova, L. et al. Genome
of _Methylobacillus flagellatus_, molecular basis for obligate methylotrophy, and polyphyletic origin of methylotrophy. _J. Bacteriol._ 189, 4020–4027 (2007). Article CAS PubMed PubMed
Central Google Scholar * Tian, J., Bryk, R., Itoh, M., Suematsu, M. & Nathan, C. Variant tricarboxylic acid cycle in _Mycobacterium tuberculosis_: identification of alpha-ketoglutarate
decarboxylase. _Proc. Natl Acad. Sci. Usa._ 102, 10670–10675 (2005). Article CAS PubMed PubMed Central ADS Google Scholar * Wagner, T., Bellinzoni, M., Wehenkel, A., O’Hare, H. M.
& Alzari, P. M. Functional plasticity and allosteric regulation of α-ketoglutarate decarboxylase in central mycobacterial metabolism. _Chem. Biol._ 18, 1011–1020 (2011). Article CAS
PubMed Google Scholar * McCloskey, D. et al. Growth adaptation of _gnd_ and _sdhCB Escherichia coli_ deletion strains diverges from a similar initial perturbation of the transcriptome.
_Front Microbiol._ 9, 1793 (2018). Article PubMed PubMed Central Google Scholar * Baba, T. et al. Construction of _Escherichia coli_ K-12 in-frame, single-gene knockout mutants: the Keio
collection. _Mol. Syst. Biol_. 2, 2006.0008 (2006). * Jiang, Y. et al. Multigene editing in the _Escherichia coli_ genome via the CRISPR-Cas9 system. _Appl. Environ. Microbiol._ 81,
2506–2514 (2015). Article CAS PubMed PubMed Central ADS Google Scholar * Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in _Escherichia coli_ K-12 using
PCR products. _Proc. Natl. Acad. Sci. USA_ 97, 6640–6645 (2000). Article CAS PubMed PubMed Central ADS Google Scholar * Saragliadis, A., Trunk, T. & Leo, J. C. Producing gene
deletions in _Escherichia coli_ by P1 transduction with excisable antibiotic resistance cassettes. _J. Vis. Exp_. 139, e58267 (2018). * Liu, B. et al. Efficient production of
3-hydroxypropionate from fatty acids feedstock in _Escherichia coli_. _Metab. Eng._ 51, 121–130 (2019). Article CAS PubMed ADS Google Scholar * Cherepanov, P. P. & Wackernagel, W.
Gene disruption in _Escherichia coli_: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. _Gene_ 158, 9–14 (1995). Article CAS PubMed
Google Scholar * Moore, S. D. Assembling new _Escherichia coli_ strains by transduction using phage P1. _Methods Mol. Biol_. 765, 155–169 (2011). * Wang, X. W. et al. GREACE-assisted
adaptive laboratory evolution in endpoint fermentation broth enhances lysine production by _Escherichia coli_. _Micro. Cell Fact._ 18, 106 (2019). Article CAS Google Scholar * Li, H. et
al. The Sequence Alignment/Map format and SAMtools. _Bioinformatics_ 25, 2078–2079 (2009). Article PubMed PubMed Central Google Scholar * Li, H. A statistical framework for SNP calling,
mutation discovery, association mapping and population genetical parameter estimation from sequencing data. _Bioinformatics_ 27, 2987–2993 (2011). Article CAS PubMed PubMed Central
Google Scholar * Long, C. P., Gonzalez, J. E., Sandoval, N. R. & Antoniewicz, M. R. Characterization of physiological responses to 22 gene knockouts in _Escherichia coli_ central carbon
metabolism. _Metab. Eng._ 37, 102–113 (2016). Article CAS PubMed PubMed Central Google Scholar * Zhang, Y. W., Zhou, H., Tao, Y. & Lin, B. X. Reconstitution of the ornithine cycle
with arginine: Glycine amidinotransferase to engineer _Escherichia coli_ into an efficient whole-cell catalyst of guanidinoacetate. _ACS Synth. Biol._ 9, 2066–2075 (2020). Article CAS
PubMed Google Scholar * Sandberg, T. E. et al. Evolution of _E. coli_ on [U-13C] glucose reveals a negligible isotopic influence on metabolism and physiology. _PLoS One_ 11, e0151130
(2016). * Long, C. P., Gonzalez, J. E., Feist, A. M., Palsson, B. O. & Antoniewicz, M. R. Fast growth phenotype of _E. coli_ K-12 from adaptive laboratory evolution does not require
intracellular flux rewiring. _Metab. Eng._ 44, 100–107 (2017). Article CAS PubMed PubMed Central Google Scholar * Long, C. P. & Antoniewicz, M. R. Quantifying biomass composition by
gas chromatography/mass spectrometry. _Anal. Chem._ 86, 9423–9427 (2014). Article CAS PubMed PubMed Central Google Scholar * Long, C. P., Au, J., Gonzalez, J. E. & Antoniewicz, M.
R. 13C metabolic flux analysis of microbial and mammalian systems is enhanced with GC-MS measurements of glycogen and RNA labeling. _Metab. Eng._ 38, 65–72 (2016). Article CAS PubMed
PubMed Central Google Scholar * McConnell, B. O. & Antoniewicz, M. R. Measuring the composition and stable-isotope labeling of algal biomass carbohydrates via gas chromatography/mass
spectrometry. _Anal. Chem._ 88, 4624–4628 (2016). Article CAS PubMed Google Scholar * Fernandez, C. A., DesRosiers, C., Previs, S. F., David, F. & Brunengraber, H. Correction of 13C
mass isotopomer distributions for natural stable isotope abundance. _J. Mass Spectrom._ 31, 255–262 (1996). Article CAS PubMed ADS Google Scholar * Antoniewicz, M. R., Kelleher, J. K.
& Stephanopoulos, G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. _Anal. Chem._ 79, 7554–7559 (2007). Article CAS PubMed Google Scholar
* Crown, S. B., Long, C. P. & Antoniewicz, M. R. Integrated 13C-metabolic flux analysis of 14 parallel labeling experiments in _Escherichia coli_. _Metab. Eng._ 28, 151–158 (2015).
Article CAS PubMed PubMed Central Google Scholar * Long, C. P., Au, J., Sandoval, N. R., Gebreselassie, N. A. & Antoniewicz, M. R. Enzyme I facilitates reverse flux from pyruvate to
phosphoenolpyruvate in _Escherichia coli_. _Nat. Commun._ 8, 14316 (2017). Article CAS PubMed PubMed Central ADS Google Scholar * Leighty, R. W. & Antoniewicz, M. R. Parallel
labeling experiments with [U-13C] glucose validate _E. coli_ metabolic network model for 13C metabolic flux analysis. _Metab. Eng._ 14, 533–541 (2012). * Yoo, H., Antoniewicz, M. R.,
Stephanopoulos, G. & Kelleher, J. K. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. _J. Biol. Chem._ 283, 20621–20627 (2008). Article CAS
PubMed PubMed Central Google Scholar * Antoniewicz, M. R., Kelleher, J. K. & Stephanopoulos, G. Elementary metabolite units (EMU): a novel framework for modeling isotopic
distributions. _Metab. Eng._ 9, 68–86 (2007). Article CAS PubMed Google Scholar * Antoniewicz, M. R. Parallel labeling experiments for pathway elucidation and 13C metabolic flux
analysis. _Curr. Opin. Biotechnol._ 36, 91–97 (2015). Article CAS PubMed Google Scholar * Antoniewicz, M. R., Kelleher, J. K. & Stephanopoulos, G. Determination of confidence
intervals of metabolic fluxes estimated from stable isotope measurements. _Metab. Eng._ 8, 324–337 (2006). Article CAS PubMed Google Scholar * Antoniewicz, M. R. et al. Metabolic flux
analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of _E. coli_ producing 1,3-propanediol. _Metab. Eng._ 9, 277–292 (2007). Article CAS PubMed PubMed
Central Google Scholar * Gonzalez, J. E., Long, C. P. & Antoniewicz, M. R. Comprehensive analysis of glucose and xylose metabolism in under aerobic and anaerobic conditions by 13C
metabolic flux analysis. _Metab. Eng._ 39, 9–18 (2017). Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by the National Key R&D Program
of China (2022YFC2106000 and 2018YFA0901400, Baixue Lin). C.P.L. and M.R.A. were supported by NSF MCB-1616332 grant. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * CAS Key Laboratory of
Microbial Physiological and Metabolic Engineering, State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, 100101, Beijing, China Hang Zhou,
Yiwen Zhang, Yanfen Xue, Yanhe Ma, Yong Tao & Baixue Lin * University of Chinese Academy of Sciences, 100049, Beijing, China Hang Zhou, Yiwen Zhang, Yong Tao & Baixue Lin *
Department of Chemical and Biomolecular Engineering, Metabolic Engineering and Systems Biology Laboratory, University of Delaware, Newark, DE, 19716, USA Christopher P. Long & Maciek R.
Antoniewicz * School of Biology and Biological Engineering, South China University of Technology, Guangzhou, 510006, China Xuesen Xia * Department of Chemical Engineering, University of
Michigan, Ann Arbor, MI, USA Maciek R. Antoniewicz Authors * Hang Zhou View author publications You can also search for this author inPubMed Google Scholar * Yiwen Zhang View author
publications You can also search for this author inPubMed Google Scholar * Christopher P. Long View author publications You can also search for this author inPubMed Google Scholar * Xuesen
Xia View author publications You can also search for this author inPubMed Google Scholar * Yanfen Xue View author publications You can also search for this author inPubMed Google Scholar *
Yanhe Ma View author publications You can also search for this author inPubMed Google Scholar * Maciek R. Antoniewicz View author publications You can also search for this author inPubMed
Google Scholar * Yong Tao View author publications You can also search for this author inPubMed Google Scholar * Baixue Lin View author publications You can also search for this author
inPubMed Google Scholar CONTRIBUTIONS H.Z., B.L. and Y.T. conceived and designed the experiments. H.Z., Y.Z., X.X. and Y.X. performed the experiments. C.P. L. and M.R.A. performed the
experiments of metabolic flux analysis. All authors contributed to data analysis. B.L., Y.T. and Y. M. supervised the study. H.Z., B.L, M.R.A, and Y.T. wrote the manuscript with
contributions from all coauthors. CORRESPONDING AUTHORS Correspondence to Yanhe Ma, Maciek R. Antoniewicz, Yong Tao or Baixue Lin. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare
no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Communications_ thanks Fabien Letisse and the other, anonymous, reviewer(s) for their contribution to the peer review of
this work. A peer review file is available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY DATA 1 SUPPLEMENTARY DATA 2 SUPPLEMENTARY DATA
3 SUPPLEMENTARY DATA 4 SUPPLEMENTARY DATA 5 SUPPLEMENTARY DATA 6 SUPPLEMENTARY DATA 7 SUPPLEMENTARY DATA 8 SUPPLEMENTARY DATA 9 SUPPLEMENTARY DATA 10 REPORTING SUMMARY SOURCE DATA SOURCE
DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and
reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes
were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If
material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain
permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Zhou, H., Zhang, Y., Long, C.P. _et al._ A citric acid cycle-deficient _Escherichia coli_ as an efficient chassis for aerobic fermentations. _Nat Commun_ 15, 2372 (2024).
https://doi.org/10.1038/s41467-024-46655-4 Download citation * Received: 21 April 2023 * Accepted: 05 March 2024 * Published: 15 March 2024 * DOI: https://doi.org/10.1038/s41467-024-46655-4
SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to
clipboard Provided by the Springer Nature SharedIt content-sharing initiative