The impact of physiological metabolite levels on serine uptake, synthesis and utilization in cancer cells
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Serine is a non-essential amino acid that is critical for tumour proliferation and depletion of circulating serine results in reduced tumour growth and increased survival in various
cancer models. While many cancer cells cultured in a standard tissue culture medium depend on exogenous serine for optimal growth, here we report that these cells are less sensitive to
serine/glycine depletion in medium containing physiological levels of metabolites. The lower requirement for exogenous serine under these culture conditions reflects both increased _de novo_
serine synthesis and the use of hypoxanthine (not present in the standard medium) to support purine synthesis. Limiting serine availability leads to increased uptake of extracellular
hypoxanthine, sparing available serine for other pathways such as glutathione synthesis. Taken together these results improve our understanding of serine metabolism in physiologically
relevant nutrient conditions and allow us to predict interventions that may enhance the therapeutic response to dietary serine/glycine limitation. SIMILAR CONTENT BEING VIEWED BY OTHERS
SERINE SYNTHESIS PATHWAY INHIBITION COOPERATES WITH DIETARY SERINE AND GLYCINE LIMITATION FOR CANCER THERAPY Article Open access 14 January 2021 THE INS AND OUTS OF SERINE AND GLYCINE
METABOLISM IN CANCER Article 28 January 2021 CAN SMALL MOLECULAR INHIBITORS THAT STOP DE NOVO SERINE SYNTHESIS BE USED IN CANCER TREATMENT? Article Open access 30 April 2021 INTRODUCTION
Numerous studies have explored the potential of dietary interventions for cancer therapy and an increasing understanding of the metabolism of cancer cells has led to the development of
therapeutic strategies focused on the dietary depletion of selected nutrients1,2. Serine, a non-essential amino-acid implicated in numerous different metabolic processes such as antioxidant
defence, one-carbon metabolism and de novo nucleotide synthesis, has been identified as a key nutrient to support tumour cell growth in vitro and in vivo3,4. Mice fed with a
serine/glycine-depleted diet show significant reduction of circulating levels of serine, leading to a reduction of tumour growth and increased survival in xenograft and genetically
engineered mouse models of cancer5,6,7,8,9. However, the sensitivity of tumours to dietary serine depletion can be modulated by additional genetic alterations. Cancers arising following
amplification of phosphoglycerate dehydrogenase (PHGDH), the first enzyme in the de novo synthesis pathway for serine, for example, can be more resistant to lack of serine in the diet while
loss of one of the serine synthesis pathway (SSP) enzymes can increase sensitivity to dietary serine depletion8. The SSP can also be activated by several common oncogenic alterations such as
activation in KRAS while loss of p53 can make tumours more sensitive to a reduction in circulating serine3,6,7,10. In light of the success of dietary intervention approaches in mouse
models, there has been an increased interest in translating this research into patients in order to improve the response to cancer therapies1,2. However, most of the in vitro studies
examining responses to nutrient deprivation have been carried out using classical cell culture media, which were formulated half a century ago with the aim of deriving and easily maintaining
mouse or human cell lines in culture11. These formulations did not attempt to recapitulate the human metabolic environment and lack some metabolites present in human circulation while
containing an over-abundance of others12. Recently, there has been increasing evidence that nutrient levels in culture media can influence the metabolic phenotype and even the response to
targeted therapies13,14. This has led to the development of culture techniques and tailored media that better represent in vivo environments such as the brain or the blood
circulation15,16,17,18. These studies have resulted in the formulation of two physiologically relevant media with similar compositions, Plasmax and HPLM16,17. In breast cancer cells, the
high level of pyruvate available in DMEM compared to Plasmax, has been shown to stabilise the hypoxia inducible factor 1a (HIF1a) leading to an hypoxic response even under normoxia16. In
addition, the high level of arginine in DMEM was shown to reverse a reaction in the urea cycle catalysed by the arginosuccinate lyase16. This response was not observed when cells were grown
in Plasmax, which contains lower, physiological levels of arginine. Untargeted metabolomics also revealed that spheroids grown in Plasmax were metabolically more similar to breast tumours
than spheroids grown in classical media16. Additionally, urate, the end product of nucleotide catabolism, is a metabolite that is present in human circulation but absent in conventional cell
culture media. Interestingly, urate present in HPLM can inhibit UMP synthetase (UMPS) and so regulate pyrimidine synthesis. UMPS is also responsible for the metabolism and cytotoxicity of
5-FU, and cell cultured in HPLM were shown to be less sensitive to 5-FU due to the presence of urate17. Another recent study using CRISPR screens to identify genetic dependencies in blood
cancer cells revealed a number of genes that were selectively essential in HPLM19. While the physiological media do not fully recapitulate the complexity of a human tumour microenvironment,
these studies show that useful information about the response of cells to metabolic interventions can be obtained using these media conditions. There has been considerable interest in the
potential for serine limitation during cancer therapy, and we have used Plasmax to more fully understand how cancer cells respond to serine limitation in patients and to identify potential
targets that could enhance the efficacy of dietary serine starvation. In this work, we observe that cells grown in Plasmax are less sensitive to serine/glycine deprivation due to a
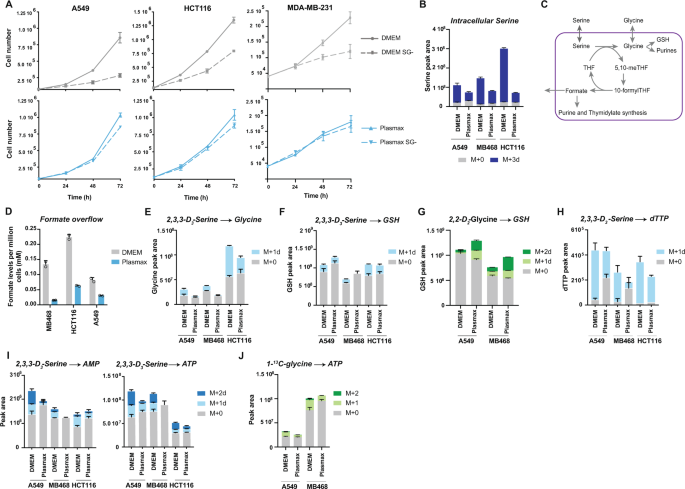
multi-layer rewiring of serine metabolism in these more physiologically relevant conditions. RESULTS SERINE AND GLYCINE METABOLISM IN PLASMAX We examined three cancer cell lines from
different cancer types, A549 (lung cancer), HCT116 (colon cancer) and MDA-MB-231 (breast cancer), whose proliferation is highly dependent on exogenous serine and glycine when grown in DMEM
(Fig. 1A)3,4. The proliferation rate of each of these cell lines was similar in DMEM or Plasmax even though Plasmax contains 4-fold less FBS than DMEM (2.5% for Plasmax and 10% for DMEM)
(Supplementary Fig. 1A). However, cells in Plasmax were clearly less sensitive to serine/glycine depletion (Fig. 1A). All the proliferation assays were carried out in media containing
non-dialysed serum (standard for these assays), which provides low levels of some nutrients, including serine. However, since Plasmax contains less serum than DMEM, this residual serine
cannot account for the difference in proliferation. For comparison, MDA-MB-468 (breast cancer) cells that carry a PHGDH amplification10 remained resistant to serine/glycine restriction in
DMEM and Plasmax (Supplementary Fig. 1B). Consistent with previous observation, extracellular serine was less rapidly taken up from Plasmax compared to DMEM without any compensatory increase
in glycine uptake (Supplementary Fig. 1C, D)16. Furthermore, increasing the serine and glycine levels in Plasmax (140 µM and 330 µM, respectively) to those found in DMEM (400 µM) did not
enhance the rate of serine uptake of A549 cells (Supplementary Fig. 1E). Plasmax and DMEM contain different concentrations of several other amino acids (Supplementary Fig. 1F), including
alanine, proline, glutamate, asparagine and aspartate which are not present in DMEM. We therefore considered whether any of these differences might influence the ability of cells to import
serine. Serine is a neutral amino acid that can be transported into the cell through the sodium-dependent transporter ASC (ASCT1 and ASCT2), the sodium-dependent transporter system A (SNAT1
and SNAT2) or the sodium-independent transporter (ASC1)20. These transporters are also able to shuttle other neutral amino acids such as alanine, which is present in Plasmax (500 µM) but not
in DMEM (0 µM)21. Alanine has been shown to inhibit L-serine transport by ASCT1 and ASCT2 when expressed in HEK cells22 and we noted that _ASCT1 (SLC1A4)_ and _ASCT2 (SLC1A5)_ transcription
is upregulated in cells grown in Plasmax (Supplementary Fig. 1G). Removal of alanine from Plasmax increased intracellular levels of labelled serine and decreased the levels of labelled
serine in the supernatant of HCT116, A549 and MDA-MB-468 (Supplementary Fig. 1H, I), while supplementing DMEM with alanine reduced the intracellular levels of serine in A549 cells
(Supplementary Fig. 1J). Taken together, these data suggest that the composition of the media can influence the transport of amino acids, such as serine. The reduced serine uptake in cells
cultured in Plasmax resulted in lower intracellular serine levels (Fig. 1B), prompting us to consider whether the utilisation of serine in these cells differed from cells grown in DMEM.
Serine is used in numerous metabolic pathways, providing glycine and one-carbon units for lipid and nucleotide synthesis and also maintaining the redox balance through glutathione and NADPH
synthesis (Fig. 1C)23. When serine is plentiful, the flux through the one-carbon cycle exceeds requirements for nucleotide synthesis and results in formate overflow24. Cells cultured in
Plasmax (for metabolomics and NMR studies dialysed serum was used to avoid confounding contributions from serum metabolites) consistently showed significantly reduced formate overflow as
measured by NMR, suggesting a lower rate of one-carbon metabolism (Fig. 1D). Using deuterium-labelled serine (2,3,3-D3-Serine), we found that there is a lower contribution of extracellular
serine to the intracellular pool of glycine in cells grown in Plasmax, as shown by reduced levels of M + 1 glycine (Fig. 1E and Supplementary Fig. 1K). The extracellular serine contribution
to the glutathione (GSH) pool in Plasmax was also slightly reduced (Fig. 1F) while cells increased the use of exogenous glycine to maintain the levels of GSH, as observed by higher labelling
from glycine into M + 2 GSH in Plasmax using deuterium labelled glycine (2,2-D2 Glycine) (Fig. 1G and Supplementary Fig. 1L). Overall, GSH levels appeared to be protected in cells grown in
Plasmax. Furthermore, there was a clear reduction in exogenous serine contribution to de novo purine and thymidylate synthesis, indicated by a lower accumulation of M + 1 dTTP and M + 2 AMP
and ATP in cells grown in the presence of deuterium-labelled serine (2,3,3-D3 Serine) (Fig. 1H, I) accompanied by a small drop in total thymidylate (in MDA-MB-468 and HCT116 cells) and total
AMP and ATP (MDA-MB-468 and A549 cells). Interestingly, these cells also show less ATP derived from carbon-labelled glycine (Fig. 1J), suggesting a reduction in de novo purine synthesis
from both extracellular serine and glycine. These data suggest that cells grown in Plasmax accommodate to decreased serine uptake by reducing one-carbon flux—thereby lowering the excess
production of formate—and channelling exogenous glycine to maintain GSH levels (Fig. 1G). The decrease in use of extracellular serine and glycine for purine and thymidylate synthesis does
not, however, strongly impact overall levels of these nucleotides and proliferation rates are maintained. UPREGULATION OF THE DE NOVO SERINE SYNTHESIS PATHWAY IN PLASMAX Our observations
that cells cultured in Plasmax generated lower amounts of purines from exogenous serine led us to examine the possibility that these cells increase de novo serine synthesis. The serine
synthesis pathway (SSP) starts with the glycolytic intermediate, 3-phosphoglycerate (3-PG), which is oxidised to 3-phosphohydroxypyruvate (3-PHP) by PHGDH, followed by two further reactions
catalysed by phosphoserine aminotransferase (PSAT1) and phosphoserine phosphatase (PSPH) to produce serine (Fig. 2A). This pathway is upregulated in serine-depleted conditions25,26 and in
some tumours, including a subset of colorectal, breast and non-small cell lung (NSCL) cancers10,27,28. Using A549 and HCT116, we found an increase in the percentage of M + 3 serine and M + 2
glycine derived from fully labelled glucose (U-13C-Glucose) in cells cultured in Plasmax (5.5 mM glucose) compared to DMEM (22.5 mM glucose) (Fig. 2B and Supplementary Fig. 2A). Increasing
the serine and glycine concentration in Plasmax to that present in DMEM did not prevent this increase in glucose-derived serine in either cell line (Supplementary Fig. 2B). While Plasmax has
lower levels of glucose than DMEM, increasing the glucose concentration in Plasmax or decreasing them in DMEM did not lead to a change in the pool of M + 3 serine (Supplementary Fig. 2C).
The enhanced de novo serine synthesis in Plasmax was accompanied by elevated expression of all three SSP enzymes, PHGDH, PSAT1 and PSPH (Fig. 2C). Each of the SSP enzymes can be induced
through activating transcription factor-4 (ATF-4), which is typically upregulated under nutrient depletion26. Indeed, we observed increased ATF-4 protein expression in cells cultured in
Plasmax as well as induction of CHOP, another downstream target of ATF-4 (Fig. 2D). This finding led us to investigate the intracellular availability of nutrients in cells grown in these
conditions. The ratios between intracellular amino acid levels in Plasmax and DMEM were broadly consistent with the ratios of extracellular amino acids levels (Supplementary Figs. 1F and
2D), with a lower level of several amino acids in Plasmax. Modulation of numerous amino acids including arginine—which is considerably lower in Plasmax (Plasmax contains 64 µM of arginine
compared to 400 µM in DMEM)—can trigger an ATF-4 response29,30. We first examined the effect of restoring the levels of several amino acids that are lower in plasmax compared to DMEM (LEU,
ILE, MET, GLN, ARG). Only the supplementation of arginine in Plasmax to the level used in DMEM reduced ATF-4 and PHGDH expression (Supplementary Fig. 2E, F) while reducing arginine in DMEM
to levels present in Plasmax, induced ATF4 and PHGDH expression (Supplementary Fig. 2G). However, increasing the serine levels in Plasmax to 400 µM (as in DMEM) did not change the expression
of ATF-4 or PHGDH (Supplementary Fig. 2F). These results indicated that the super-physiological levels of amino acids such as arginine in DMEM suppress a level of ATF-4 activity that is
seen under more physiological concentrations of these amino acids, which is in line with upregulation of the ATF-4 pathway in human and mouse tumour compared to normal tissue31. Confirming
the increased contribution of the SSP in cells grown in Plasmax compared to DMEM, we noted an increase in the pools of M + 6, M + 7, M + 8, M + 9 labelled ATP, which appear when
U-13C-glucose-derived glycine and one-carbon units are used to synthesise purines in cells fed carbon-labelled glucose (Fig. 2E). To assess the contribution of this increase in de novo
serine synthesis in Plasmax to proliferation, we examined the effect of treating cells with the PHGDH inhibitor, PH75532. While PHGDH inhibition had no effect on cancer cells growth in DMEM,
it significantly reduced the proliferation of cells cultured in Plasmax (Fig. 2F). These data suggest that cells cultured in Plasmax are more dependent on de novo synthesised serine as a
source of one-carbon units and to support purine synthesis and proliferation. However, the total pool of intracellular serine does not significantly change after treatment with PH755
indicating that cells can revert to serine uptake during PHGDH inhibition (Supplementary Fig. 2H). This was confirmed by the rescue of the anti-proliferative effect of PH755 by supplementing
the cells with excess serine (Supplementary Fig. 2I). In addition, while removal of serine and glycine from either DMEM or Plasmax led to an increase in de novo serine synthesis, this
response was more robust in Plasmax, where this pathway was already induced in the presence of exogenous serine (Fig. 2G). It would appear that amino acid availability increases SSP activity
in Plasmax, allowing cells to be less sensitive to the removal of exogenous serine. HYPOXANTHINE SALVAGE IS A MAJOR CONTRIBUTOR OF PURINES IN PLASMAX We next considered that other
differences between DMEM and Plasmax could impact the dependence on exogenous serine. Previous studies showed that formate, which is present at 33 µM in Plasmax but absent from DMEM, can
provide one-carbon units to support purine synthesis and—in cooperation with glycine—proliferation4. Tracing 13C-labelled formate into ATP in both Plasmax and DMEM showed that cells grown in
either media could access exogenous formate for purine synthesis (Supplementary Fig. 3A). However, depleting formate from Plasmax did not lower the overall levels of ATP or lead to an
increased incorporation of serine-derived nitrogen and carbons into ATP (Supplementary Fig. 3B), indicating that while the free formate in Plasmax could contribute to ATP synthesis, it does
not influence the contribution of extracellular serine to de novo purine synthesis (Supplementary Fig. 3B). In addition to de novo synthesis, purines can be synthesised through the salvage
pathway, which recycles the hypoxanthine derived from degraded bases and transfers it to a phosphoribosyl group to form inosine monophosphate (IMP), so maintaining the levels of
adenylates33,34. Hypoxanthine is not present in DMEM but Plasmax contains 5 µM hypoxanthine, reflecting the level in human circulation. Cells grown in Plasmax rapidly deplete hypoxanthine
from the extracellular media and accumulate high intracellular levels compared to cells grown in DMEM (Fig. 3A, B). To determine whether the lower uptake of exogenous serine seen in cells
grown in Plasmax is the result of hypoxanthine utilisation, we removed hypoxanthine from the Plasmax and looked at incorporation of fully labelled 13C3-15N-serine into ATP. As observed in
Fig. 1I, exogenous serine incorporation into ATP was lower in the three cells lines cultured in Plasmax compared to DMEM (Fig. 3C). However, removal of hypoxanthine from Plasmax fully
restored the levels of exogenous serine utilisation to those seen in DMEM (Fig. 3C). Furthermore, addition of hypoxanthine to DMEM significantly reduced the incorporation of serine into ATP,
confirming that cancer cells preferentially salvage hypoxanthine instead of synthesising de novo ATP from extracellular serine (Supplementary Fig. 3C). Indeed, by tracing deuterium-labelled
hypoxanthine we evaluated its direct contribution to the ATP pool, which consistently ranged from 20 to 40% in both DMEM and Plasmax (Fig. 3D and Supplementary Fig. 3D). This proportion of
ATP derived from hypoxanthine is generally higher than the pools of ATP synthesised from media-derived or de novo synthesised serine (Supplementary Fig. 3E). Furthermore, removal of
hypoxanthine from Plasmax increased the proportions of de novo synthesised serine labelling of ATP, along with an increase in ATP derived from exogenous serine (Fig. 3E and Supplementary
Fig. 3E). Taken together, these data suggest that both de novo serine and hypoxanthine contribute to the pools of ATP in cells cultured in Plasmax. Our findings corroborate different studies
in human cancer patients that suggest that cancer cells do not exclusively synthetise purines from extracellular serine but partially from hypoxanthine35,36,37. In the cell lines that we
have tested, hypoxanthine salvage seems to be preferentially used to provide purines instead of de novo synthesis from serine. Next, we aimed to determine if the presence of hypoxanthine was
able to mitigate the anti-proliferative effect of serine and glycine deprivation in physiological conditions. To assess this in vitro and in vivo, we included DLD-1, a colorectal cancer
cell line resistant to serine/glycine starvation in DMEM and which we have previously studied in vivo9. When cultured in Plasmax, we observed that DLD-1 cells showed enhanced de novo serine
synthesis, became sensitive to PHGDH inhibition with PH755 and had a reduced contribution of serine to de novo purine synthesis due to the presence of hypoxanthine, similar to the other cell
lines (Supplementary Fig. 3F–H). COOPERATION BETWEEN SERINE LIMITATION AND MODULATION OF HYPOXANTHINE IN LIMITING TUMOUR CELL GROWTH To test the contribution of hypoxanthine to purine pools
when serine availability is limited, we first measured intracellular levels of labelled hypoxanthine when cells were grown in Plasmax and treated with the PHGDH inhibitor or serine and
glycine deprivation. We observed increased levels of intracellular hypoxanthine both after serine/glycine starvation and PHGDH inhibition (Fig. 3F and Supplementary Fig. 3I). Extracellular
levels of hypoxanthine were also depleted after treatment with PH755 (Fig. 3G). In addition, even though hypoxanthine supplementation in full medium did not affect cell proliferation
(Supplementary Fig. 4A), hypoxanthine supplementation rescued proliferation of cells in serine/glycine-free DMEM, presumably by allowing endogenously produced serine and glycine to be used
in processes other than de novo purine synthesis (Fig. 4A). The hypoxanthine also partially rescued proliferation in serine-starved cells supplemented with glycine, similar to the effect of
formate (Fig. 4B)38 and supporting the proposal that cells use hypoxanthine to maintain ATP levels. Cell proliferation in serine/glycine-free Plasmax depleted of hypoxanthine was only
modestly reduced (Supplementary Fig. 4B). However, combination of PH755 with a lack of hypoxanthine resulted in further reduction of cancer cell proliferation compared to treatment of PH755
alone (Fig. 4C). These results support the increased dependence of cells in Plasmax on endogenous serine synthesis and indicate that the increased hypoxanthine uptake in response to PHGDH
inhibition can support cell proliferation. Tumours are irrigated by the vasculature, which provides the interstitial space with a nutrient rich fluid termed the interstitial fluid (TIF)39.
As shown previously, we found that circulating hypoxanthine levels in mice (0.02–0.2 µM) were lower than those in humans (∼5 µM)17. Furthermore, TIF from tumours formed from DLD-1
cells—which utilised hypoxanthine in vitro—also showed a similarly low range of hypoxanthine levels (∼0.18 µM for plasma and ∼0.77 µM for TIF (Supplementary Fig. 4C). To determine the impact
of serine/glycine limitation on circulating and tumour levels of hypoxanthine, we analysed serum and tumours from mice carrying DLD-1 xenograft tumours and treated with a
serine/glycine-free diet in combination with the PHGDH inhibitor, PH755. These samples were generated for a previous study that showed an inhibition of tumour growth in the double treated
mice9. Although there was considerable variability between samples, we observed a trend towards reduction in circulating hypoxanthine levels (Fig. 4D) that paralleled the reduction in
circulating serine and glycine levels (Supplementary Fig. 4D). Interestingly, tumoral levels of hypoxanthine did not decrease, consistent with an ability of tumour cells to take up exogenous
hypoxanthine to support nucleotide synthesis under serine limiting conditions (Fig. 4E). Our work predicts that limiting hypoxanthine metabolism would cooperate with limitation of serine
availability to inhibit the growth of cancer cells in patients. This may be difficult to test in mice, where the concentration of circulating hypoxanthine (0.2 uM) was shown to be
insufficient to improve the proliferation of serine/glycine starved cells (Fig. 4A, B). We therefore looked for a potential cooperation between serine starvation and hypoxanthine limitation
in Plasmax. Initially we treated cells with 6-mercaptopurine (6-MP), an anti-metabolite that competes with hypoxanthine for hypoxanthine-guanine phosphoribosyltransferase (HPRT), the enzyme
required to utilise hypoxanthine for purine synthesis. HPRT activity also leads to the conversion of 6-MP into 5-thio-ionsinic acid (TIMP), which inhibits the conversion of IMP to AMP. Upon
treatment with 6-MP for 2 h we detected the expected production of TIMP (Supplementary Fig. 4E) and some inhibition of proliferation after 72 h (Fig. 4F) although this treatment did not
affect the proliferation of cells in DMEM (Supplementary Fig. 4F). The combination of 6-MP and PH755 resulted in a significantly greater decrease in proliferation compared to each single
treatment in 3 out of 4 cell lines (Fig. 4F). To assess directly the effect of a reduction in HPRT activity, we used siRNA to deplete cells of HPRT expression (Supplementary Fig. 4G).
Knockdown of HPRT led to a reduction of cell growth in Plasmax confirming the importance of this pathway in physiological medium (Fig. 4G). Interestingly, although removal of hypoxanthine
from Plasmax leads to increased purine production from both de novo synthesised serine and exogenous serine (Supplementary Fig. 3E), the combination of HPRT depletion and PHGDH inhibition
resulted in complete abrogation of cell proliferation (Fig. 4G). Since these cells show enhanced serine uptake, these results suggest additional consequences of HPRT depletion leading to
proliferative arrest. Taken together, these observations show that the resistance of cells in Plasmax to serine and glycine deprivation is due to a combination of mechanisms reflecting the
rewiring of the serine and one-carbon metabolism in this culture medium. DISCUSSION Our data indicate that the contribution of de novo serine synthesis in cancer cells to the nucleotide pool
is underestimated when cells are cultured in classical growth media—in which a greater dependency on exogenous serine is seen. Using Plasmax to more accurately represent metabolite levels
available in human plasma, we noted a higher basal level of SSP activity compared to classical media reflected in elevated ATF-4 expression, potentially reflecting levels of amino-acids such
as arginine. The higher SSP enzyme expression and activity that we observe in physiological media is in line with the increasing evidence that SSP enzymes are highly important in cancer
biology6,40,41. This study also demonstrates the contribution that hypoxanthine provides to support purine synthesis and reduce the dependence on exogenous serine and glycine. Previous
studies have suggested that neoplastic cells initiate increased purine salvage to gain a growth advantage36, with elevation of the expression of HPRT, a key enzyme in the salvage pathway, in
50% of lung carcinoma patients42. Although hypoxanthine was reduced in the plasma of tumour bearing mice fed with a serine/glycine-deprived chow and treated with a de novo serine synthesis
inhibitor, maintenance of hypoxanthine levels in the tumour suggest that this metabolite is available and utilised by the tumour when serine is limiting. While dietary strategies to limit
circulating serine and glycine have been shown to be therapeutically effective in a number of mouse cancer models, this work suggests that targeting de novo serine synthesis and hypoxanthine
availability may significantly enhance the response to circulating serine and glycine limitation. Dietary modulation of hypoxanthine has been described43 and the targeting of hypoxanthine
transporters can affect tumour proliferation44. Our study highlights the potential of physiological media to reveal metabolic vulnerabilities of cancer cells that may provide additional
therapeutic targets. METHODS CELL CULTURE All cell lines were obtained from the Francis Crick Institute Cell Services Science Technology Platform, which ensures various quality controls such
as mycoplasma testing, STR profiling and species validation. All cell lines were maintained in DMEM (Gibco, 41966) supplemented with 10% FBS and penicillin–streptomycin at 37 °C in
humidified atmosphere of 5% CO2. For experimental work, cells were transferred to test media 24 h prior to the start of the experiments. Test media was formulated with MEM (Gibco)
supplemented with 2 mM L-glutamine, 17 mM D-glucose (to a final concentration of 22.5 mM glucose) and MEM vitamins (SG- medium) while control media was further supplemented with 400 µM
L-serine and glycine (termed DMEM). Plasmax and all its formulation in the manuscript were made in the lab on the basis of the formulation previously published16. PROLIFERATION ASSAY For all
proliferation assays non-dialysed FBS was used at 10% in DMEM and 2.5% in Plasmax. For Fig. 1A, Supplementary Fig. 1A and Supplementary Fig. 1B all cell lines were plated at 2 × 104 cells
in 6-well plates in DMEM with 3 replicates per time point. After 24 h cells were washed with PBS and transferred to the experimental conditions. Cells were then trypsinized, diluted in
PBS-EDTA and counted with a CASY Model TT Cell Counter (Innovatis, Roche Applied Science). For Fig. 2F and Supplementary Fig. 3F, A549, HCT116, MDA-MB-468 and DLD-1 cells were plated at 2.5
× 104 cells in a 12-well plate in DMEM with three replicates per time point and condition. After 24 h of acclimatisation in Plasmax or DMEM cells were washed with PBS and transferred to the
experimental conditions and treated with 10 µM PH755 (Raze Therapeutics) diluted in DMSO or DMSO alone. Media were changed after 48 h and cells were counted as described above. For
Supplementary Fig. 2I, HCT116, A549 and MDA-MB-468 were plated at 2.5 × 104 cells in a 12-well plate in Plasmax with three replicates per condition. After 24 h of acclimatisation in Plasmax
cells were washed with PBS and transferred to the different experimental conditions. Where indicated cells were treated with 10 µM PH755 (Raze Therapeutics) diluted in DMSO or DMSO alone.
For serine supplementation, cells in Plasmax received an additional 400 µM diluted in PBS. Media were changed after 48 h and cells were counted as described above. Relative cell number was
calculated to the number of cells after 72 h in the complete Plasmax media condition. In Supplementary Fig. 4A, A549 and HCT116 cells were plated at 5 × 104 cells in a 12-well plate in DMEM
with three replicates per time point and condition. In Fig. 4A, B, A549, HCT116 and DLD-1 cells were plated at 5 × 104 in a 12-well plate in DMEM with three replicates per time point and
condition. After 24 h of acclimatisation in Plasmax or DMEM cells in indicated media was supplemented with 5 µM or 0.2 µM hypoxanthine diluted in DMSO or DMSO alone from time point 0. Cell
counting was performed as described above. Media were changed after 48 h and cells were counted as described above. For Fig. 4C and Supplementary Fig. 4B, HCT116, A549, MDA-MB-468 and DLD-1
were plated at 2.5 × 104 cells in a 12-well plate in Plasmax with three replicates per condition. After 24 h of acclimatisation in Plasmax cells were washed with PBS and transferred to the
different experimental conditions. For Fig. 4C cells received 10 µM of PH755 (Raze Therapeutics) diluted in DMSO or DMSO alone when indicated. Media were changed after 48 h and cells were
counted as described above after 72 h. Relative cell number was calculated to the number of cells after 72 h in the complete Plasmax media condition. For Fig. 4F, HCT116, A549, MDA-MB-468
and DLD-1 were plated at 2.5 × 104 cells in a 12-well plate in Plasmax with three replicates per condition. After 24 h of acclimatisation in Plasmax cells were washed with PBS and
transferred to the different experimental conditions. When indicated cells received 10 µM of PH755 (Raze Therapeutics) diluted in DMSO or DMSO alone. For 6-Mercaptopurine (6-MP) (Merck)
MDA-MB-468 cells received 1 µM, A549 and HCT116 received 2.5 µM and DLD-1 received 5 µM diluted in DMSO when indicated. Media were changed after 48 h and cells were counted as described
above after 72 h. Relative cell number was calculated to the number of cells after 72 h in the complete Plasmax media condition. For Fig. 4G, HCT116, A549, MDA-MB-468 and DLD-1 were plated
at 2.5 × 104 cells in a 12-well plate and transfected with a pool of siRNA targeting HPRT (D-008735-01) or a non-targeting siRNA (D-001206-13) from Dharmacon. After 24 h, media were changed
to the treatment media. When indicated cells received 10 µM of PH755 (Raze Therapeutics) diluted in DMSO or DMSO alone. Media were changed after 48 h and cells were counted at 72 h. Relative
cell number was calculated to the number of cells when media were changed to treatment media. WESTERN BLOT After pelleting, cells were lysed with RIPA-buffer (Millipore) supplemented with
phosphatase inhibitor cocktail (ThermoFisher Scientific) and complete protease inhibitors (Roche). Proteins were separated using precast 4–12% bis-tris gels (Invitrogen) and transferred to
nitrocellulose by dry-transfer (iBlot, Thermofisher Scientific). Membranes were probed with the following primary antibodies: PHGDH (#13428), ATF4 (#11815) and CHOP (#2895) from Cell
Signalling Technology; PSAT (ab96136), PSPH (ab96414) and HPRT (ab133242), Actin (ab8226) from Abcam; Vinculin (sc-73614) from Santa Cruz Biotechnology. All antibodies were used at a
dilution of 1:1000. Blot were developed with ECL chemiluminescence kits (Pierce) after incubation with appropriate species-specific horseradish peroxidase-conjugated antibodies. All
antibodies were verified and confirmed for species as per the manufacturers’ disclosure. Additional validation was performed for the PHGDH antibody using a PHGDH KO cell line. PSAT and PSPH
antibodies were validated under serine/glycine free conditions. LIQUID CHROMATOGRAPHY–MASS SPECTROMETRY For all LC–MS experiments dialysed FBS was used for DMEM (10%) or Plasmax (2.5%). For
metabolites in supernatants, 2 × 105 of HCT116, A549 and MDA-MB-468 were plated in 6-well plates in DMEM for 24 h in order to attach. Cells were then switched to fresh DMEM, Plasmax, Plasmax
(400 µM SER, 400 µM GLY) or Plasmax without alanine and kept for up to 96 h. 10 µl of supernatant of each time point was extracted with 490 µL of ice-cold extraction buffer
(methanol/acetonitrile/water in the 50/30/20 ratio), vortexed and spun at 15,000 _g_ for 12 min at 4 °C. Supernatants were used for LC–MS analysis. For labelling experiments, 2 × 105 of
HCT116, A549, MDA-MB-468 and DLD-1 cells were plated in 6-well plates in the experimental media 24 h prior to the labelling. Duplicate plates were used for cell counting in order to
normalise the LC–MS analysis on the basis of cell number. After 24 h in the experimental media, media were replaced by the equivalent media depleted of the metabolite of interest and
supplemented with the same amount of the labelled metabolite for 4 h. In the case of treatment the compound was added at the same time as the labelling media. Labelled compounds used were:
13C3-15N-serine (#608130) and 2,2-D2-glycine (#336459) from Sigma-Aldrich; U-13C-glucose (CLM-1396), 1-13C-glycine (CLM-422), 2,3,3-D3-serine (DLM-582), 2,8,9-D3-hypoxanthine (DLM-2923) from
Cambridge Isotopes. Cells were washed with PBS and metabolites were extracted in the plate with the same extraction buffer as described above. After vortexing and centrifugation at 15,000
_g_ for 12 min at 4 °C, supernatants were used for LC–MS analysis. For the analysis of tumour samples, tissue (20–40 mg/mL of described extraction buffer) was homogenised in a precellys 24
homogeniser (Bertin Instruments). Homogenate was collected and spun at 15,000 _g_ for 12 min at 4 °C and the supernatant collected for LC–MS analysis. For plasma samples, plasma was diluted
20-fold with the same extraction buffer, vortexted and spun at 15,000 _g_ for 12 min at 4 °C. The supernatant was collected for analysis. For tumour interstitial fluid (TIF), extracted
tumours were placed in Spin-x filter tubes (Costar 8170) and centrifuged sequentially at 100 _g_, 400 _g_ and 1500 _g_ for 10 min at 4 °C. TIF was the diluted 20-fold in the same extraction
buffer, vortexed and spun at 15,000 _g_ for 12 min at 4 °C. Supernatant was collected for analysis. LC–MS analysis was performed as previously described4. Data were analysed with TraceFinder
and Xcalibur from Thermofisher. NMR NMR was used to measure formate as represented in Fig. 1D. HCT116, A549 and MDA-MB-468 were plated at 1 × 105 in 6 well plates and left for three days in
DMEM or Plasmax. 160 µL of supernatant was collected for NMR analysis and cells were counted as already described to allow normalisation to cell number. A solution of 10 mM DSS (sodium
2,2-dimethyl-2-silapentane-5-sulfonate from Sigma) diluted in D20 was added to the samples to obtain a final concentration of 0.1 mM DSS. NMR spectra were acquired at 25 °C with a Bruker
Avance III HD instrument with a nominal 1H frequency of 700 MHz using 3 mm tubes in a 5 mm CPTCI cryoprobe. For 1H 1D profiling spectra the Bruker pulse program _zgesgppe_ for excitation
sculpting with pure echo45 was used with 20 ppm sweep width, 1 s relaxation delay and 4 s acquisition time. Typically, 128 transients were acquired. Data were processed and analysed using
_Chenomx NMR Suite_ (Chenomx, Edmonton, Canada). Free induction decays were zero-filled, apodized with exponential multiplication (line-broadening factor LB = 1 Hz), Fourier-transformed and
the resulting spectra were then phase corrected before baseline correction, all in the Processor component of the Chenomx software. Formate quantitation was performed based on the chemical
shift reference (DSS) assumed to be at 0.1 mM concentration and with linewidth adjusted to obtain a good fit to the Chenomx library spectra for multiple metabolites in the spectrum. RT-QPCR
Total RNA was extracted using TriPure isolation reagent (Roche) from at least two technical replicates. cDNA was generated using the high-capacity cDNA reverse transcription kit
(ThermoFisher) according to manufacturer’s instructions. IDT PrimeTime® gene expression master mix (IDT) was used to perform qPCR with the following predesigned primer/probe assay from IDT;
_ASCT1_ (Hs.PT.58.4102571), _ASCT2_ (Hs.PT.58.21358468), _POLRA_ (Hs.PT.39a.19639531). Sequences can be found in Supplementary Table 1. Data were collected on a QuantStudio 3 system from
applied biosystems. IN VIVO EXPERIMENTS All experiments were conducted in compliance with the UK Home Office-approved project licences and personal licences (Animals Scientific Procedures
Act 1986) and within institutional welfare guidelines (Francis Crick Institute). Animal experiments were subject to ethical review by the Francis Crick Animal Welfare and Ethical Review Body
and carried out under UK Home Office project license P319AE968. Mice (3–5 per cage) had ad libitum access to food and water and were kept in a 12 h day/night cycle (7:00 to 19:00) in rooms
at 21 °C at 55% humidity. Mice were acclimatised to their environment for at least one week prior to experimentation. Mice used in the experiments were littermates randomly assigned to
experimental groups. The experimental diets used in Fig. 4F (control diet and –SG diet) were previously described as “Diet 1-control” and “Diet 1-SG-free”7. Control diet contains all
essential amino acids in addition to serine, glycine, glutamine, arginine, cysteine and tyrosine. The SG- diet is formulated as the control diet lacking serine and glycine which is
compensated by a proportionally increased level of the other amino acids to reach the same total amino acid content. XENOGRAFT EXPERIMENTS In Fig. 4D, E and Supplementary Fig. 4D, CD-1 nude
mice (obtained from Charles River, 7-9 weeks old) received a subcutaneous injection of 4 × 106 DLD-1 cells suspended in PBS. Mice were placed on experimental diet two days after tumour
injections. A further two days after diet change, mice were treated either with vehicle (0.5% methylcellulose (Sigma, H7509), 0.5% Tween-80 (Sigma, P8192) or PH755 (obtained by Raze
Therapeutics) prepared in vehicle once daily by oral gavage. The starting dosage of PH755 was 100 mg/kg (for seven days) and was subsequently lowered to 50 mg/kg (for six days) and increased
again to 75 mg/kg (for seven days). This experiment was generated for a previously published experiment9. In Supplementary Fig. 4C, NOD.C-PrkdcSCID Il2rgtm1Wjl/SzJ (NSG) mice in between 8
and 12 weeks old (obtained from The Francis Crick Institute breeding facility) received subcutaneous injection of 4 × 106 DLD-1 cells suspended in PBS and left to grow for 4 weeks before
harvesting the tumours. STATISTICAL ANALYSIS All statistical analyses were performed using GraphPad Prism 8 software. Unpaired Student’s _t_ test was performed to compare two groups to each
other. If the variance, determined by the F test, between the two groups was unequal, a Welch’s correction was applied. For multiple comparison, a one-way ANOVA was used. If the variance
between groups, determined by the Brown–Forsythe test, was unequal, a Brown–Forsythe and Welch correction was applied. _p_ value below 0.05 was considered statistically significant.
Significance is indicated as follows: *_p_ < 0.05, **_p_ < 0.01, ***_p_ < 0.001, ****_p_ < 0.0001, ns no significance. All measurements were taken from distinct samples, as noted
in figure legends, and no data were excluded. Sample sizes were based in standard protocols in the field, metabolic data were assigned in a random order before analysis by LC–MS, mice were
randomly assigned to a treatment and mouse experiments were blinded to the person performing metabolomic analysis. REPORTING SUMMARY Further information on research design is available in
the Nature Research Reporting Summary linked to this article. DATA AVAILABILITY All the data (including source data) supporting the findings of this study are available within the article,
the supplementary information files and the source data file. Source data are provided with this paper. REFERENCES * Kanarek, N., Petrova, B. & Sabatini, D. M. Dietary modifications for
enhanced cancer therapy. _Nature_ 579, 507–517 (2020). Article ADS CAS Google Scholar * Tajan, M. & Vousden, K. H. Dietary approaches to cancer therapy. _Cancer Cell_ 37, 767–785
(2020). Article CAS Google Scholar * Maddocks, O. D. et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. _Nature_ 493, 542–546 (2013). Article
ADS CAS Google Scholar * Labuschagne, C. F. et al. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. _Cell Rep._ 7, 1248–1258 (2014). Article
CAS Google Scholar * Gravel, S. P. et al. Serine deprivation enhances antineoplastic activity of biguanides. _Cancer Res._ 74, 7521–7533 (2014). Article CAS Google Scholar * Sullivan,
M. R. et al. Increased serine synthesis provides an advantage for tumors arising in tissues where serine levels are limiting. _Cell Metab._ 29, 1410–1421 (2019). e4. Article CAS Google
Scholar * Maddocks, O. D. K. et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. _Nature_ 544, 372–376 (2017). Article ADS CAS Google Scholar
* Banh, R. S. et al. Neurons release serine to support mRNA translation in pancreatic cancer. _Cell_ 183, 1202–1218 (2020). e25. Article CAS Google Scholar * Tajan, M. et al. Serine
synthesis pathway inhibition cooperates with dietary serine and glycine limitation for cancer therapy. _Nat. Commun._ 12, 366 (2021). Article CAS Google Scholar * Possemato, R. et al.
Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. _Nature_ 476, 346–350 (2011). Article ADS CAS Google Scholar * Lagziel, S., Gottlieb, E. &
Shlomi, T. Mind your media. _Nat. Metab._ 2, 1369–1372 (2020). Article Google Scholar * Ackermann, T. & Tardito, S. Cell culture medium formulation and its implications in cancer
metabolism. _Trends Cancer_ 5, 329–332 (2019). Article CAS Google Scholar * Muir A. et al. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase
inhibition. _Elife_ 6, e27713 (2017). * Gui, D. Y. et al. Environment dictates dependence on mitochondrial complex I for NAD+ and aspartate production and determines cancer cell sensitivity
to metformin. _Cell Metab._ 24, 716–727 (2016). Article CAS Google Scholar * Bardy, C. et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in
vitro. _Proc. Natl Acad. Sci. USA_ 112, E2725–E2734 (2015). Article ADS CAS Google Scholar * Vande Voorde, J. et al. Improving the metabolic fidelity of cancer models with a
physiological cell culture medium. _Sci. Adv._ 5, eaau7314 (2019). Article ADS Google Scholar * Cantor, J. R. et al. Physiologic medium rewires cellular metabolism and reveals uric acid
as an endogenous inhibitor of UMP synthase. _Cell_ 169, 258–272 e17 (2017). Article CAS Google Scholar * Birsoy, K. et al. Metabolic determinants of cancer cell sensitivity to glucose
limitation and biguanides. _Nature_ 508, 108–112 (2014). Article ADS CAS Google Scholar * Nicholas J. Rossiter et al. CRISPR screens in physiologic medium reveal conditionally essential
genes in human cells. _Cell Metab._ 33, 1248–1263 (2021). * Broer, S. & Broer, A. Amino acid homeostasis and signalling in mammalian cells and organisms. _Biochem. J._ 474, 1935–1963
(2017). Article CAS Google Scholar * El-Hattab, A. W. Serine biosynthesis and transport defects. _Mol. Genet. Metab._ 118, 153–159 (2016). Article CAS Google Scholar * Foster, A. C. et
al. D-Serine is a substrate for neutral amino acid transporters ASCT1/SLC1A4 and ASCT2/SLC1A5, and is transported by both subtypes in rat hippocampal astrocyte cultures. _PLoS ONE_ 11,
e0156551 (2016). Article Google Scholar * Locasale, J. W. Serine, glycine and one-carbon units: cancer metabolism in full circle. _Nat. Rev. Cancer_ 13, 572–583 (2013). Article CAS
Google Scholar * Meiser, J. et al. Serine one-carbon catabolism with formate overflow. _Sci. Adv._ 2, e1601273 (2016). Article ADS Google Scholar * Ye, J. et al. Pyruvate kinase M2
promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. _Proc. Natl Acad. Sci. USA_ 109, 6904–6909 (2012). Article ADS CAS Google Scholar * DeNicola, G. M.
et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. _Nat. Genet._ 47, 1475–1481 (2015). Article CAS Google Scholar * Jia, X. Q. et al. Increased expression of PHGDH
and prognostic significance in colorectal cancer. _Transl. Oncol._ 9, 191–196 (2016). Article Google Scholar * Zhu, J. et al. High expression of PHGDH predicts poor prognosis in non-small
cell lung cancer. _Transl. Oncol._ 9, 592–599 (2016). Article Google Scholar * Sahu, D. et al. Argininosuccinate Synthetase 1 loss in invasive bladder cancer regulates survival through
general control nonderepressible 2 kinase-mediated eukaryotic initiation factor 2alpha activity and is targetable by pegylated arginine deiminase. _Am. J. Pathol._ 187, 200–213 (2017).
Article CAS Google Scholar * Lee, J. et al. Translational control of inducible nitric oxide synthase expression by arginine can explain the arginine paradox. _Proc. Natl Acad. Sci. USA_
100, 4843–4848 (2003). Article ADS CAS Google Scholar * Ye, J. et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation.
_EMBO J._ 29, 2082–2096 (2010). Article CAS Google Scholar * Rodriguez, A. E. et al. Serine metabolism supports macrophage IL-1beta production. _Cell Metab._ 29, 1003–1011 (2019). e4.
Article CAS Google Scholar * Yin, J. et al. Potential mechanisms connecting purine metabolism and cancer therapy. _Front. Immunol._ 9, 1697 (2018). Article Google Scholar * Lane, A. N.
& Fan, T. W. Regulation of mammalian nucleotide metabolism and biosynthesis. _Nucleic Acids Res._ 43, 2466–2485 (2015). Article CAS Google Scholar * Fan, T. W. M. et al. De novo
synthesis of serine and glycine fuels purine nucleotide biosynthesis in human lung cancer tissues. _J. Biol. Chem._ 294, 13464–13477 (2019). Article CAS Google Scholar * Ong, E. S. et al.
Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. _Mol Cell Proteom._ https://doi.org/10.1074/mcp.m900551-mcp200 (2010). * Qi, Z. & Voit,
E. O. Identification of cancer mechanisms through computational systems modeling. _Transl. Cancer Res._ 3, 233–242 (2014). CAS PubMed PubMed Central Google Scholar * Diehl, F. F. et al.
Cellular redox state constrains serine synthesis and nucleotide production to impact cell proliferation. _Nat. Metab._ 1, 861–867 (2019). Article CAS Google Scholar * Sullivan M. R. et
al. Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. _Elife_ 8, e44235 (2019). * Méndez-Lucas, A. et al. Identifying
strategies to target the metabolic flexibility of tumours. _Nat. Metab._ 2, 335–350 (2020). Article Google Scholar * Ngo B. et al. Limited environmental serine and glycine confer brain
metastasis sensitivity to PHGDH inhibition. _Cancer Discov._ 10, 1352–1373 (2020). * Townsend, M. H. et al. Non-small-cell lung cancer cell lines A549 and NCI-H460 express hypoxanthine
guanine phosphoribosyltransferase on the plasma membrane. _Onco Targets Ther._ 10, 1921–1932 (2017). Article CAS Google Scholar * Zgaga, L. et al. The association of dietary intake of
purine-rich vegetables, sugar-sweetened beverages and dairy with plasma urate, in a cross-sectional study. _PLoS ONE_ 7, e38123 (2012). Article ADS CAS Google Scholar * Spano, D. et al.
Dipyridamole prevents triple-negative breast-cancer progression. _Clin. Exp. Metastasis_ 30, 47–68 (2013). Article CAS Google Scholar * Adams, R. W. et al. “Perfecting” WATERGATE: clean
proton NMR spectra from aqueous solution. _Chem. Commun._ 49, 358–360 (2013). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We would like to thank Dr. S. Tardito and Dr.
J. Van de Voorde from the Beatson Institute for providing us with the Plasmax formulation ahead of publication. Metabolomic analysis was supported by Dr. James Macrae and the metabolomics
STP of the Francis Crick Institute and through provision of access to the MRC Biomedical NMR Centre. The work was funded by Cancer Research UK grant C596/A26855 and supported by the Francis
Crick Institute which receives its core funding from Cancer Research UK (FC001557 and FC001029), the UK Medical Research Council (FC001557 and FC001029) and the Wellcome Trust (FC001557 and
FC001029). MH is in receipt of a Wallonie-Bruxelles International Grant of Excellence WBI.WORLD (SUB/2019/441394). AUTHOR INFORMATION Author notes * These authors contributed equally: Marc
Hennequart, Christiaan F. Labuschagne. AUTHORS AND AFFILIATIONS * The Francis Crick Institute, 1 Midland Road, London, NW1 1AT, UK Marc Hennequart, Christiaan F. Labuschagne, Mylène Tajan,
Steven E. Pilley, Eric C. Cheung, Nathalie M. Legrave, Paul C. Driscoll & Karen H. Vousden Authors * Marc Hennequart View author publications You can also search for this author inPubMed
Google Scholar * Christiaan F. Labuschagne View author publications You can also search for this author inPubMed Google Scholar * Mylène Tajan View author publications You can also search
for this author inPubMed Google Scholar * Steven E. Pilley View author publications You can also search for this author inPubMed Google Scholar * Eric C. Cheung View author publications You
can also search for this author inPubMed Google Scholar * Nathalie M. Legrave View author publications You can also search for this author inPubMed Google Scholar * Paul C. Driscoll View
author publications You can also search for this author inPubMed Google Scholar * Karen H. Vousden View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS The study was conceived and designed by M.H., C.L. and K.H.V. Investigations were conducted by M.H., C.L., M.T., S.P., N.L. and E.C. NMR analysis was carried out by P.D. The
manuscript was written by M.H., C.L. and K.H.V. CORRESPONDING AUTHOR Correspondence to Karen H. Vousden. ETHICS DECLARATIONS COMPETING INTERESTS KHV is on the Board of Directors and
shareholder of Bristol Myers Squibb and on the Science Advisory Board (with stock options) of PMV Pharma, RAZE Therapeutics and Volastra Therapeutics. She is also on the SAB of Ludwig
Cancer. KHV is a co-founder and consultant of Faeth Therapeutics, funded by Khosla Ventures. She has been in receipt of research funding from Astex Pharmaceuticals and AstraZeneca and
contributed to CRUK Cancer Research Technology filing of Patent Application WO/2017/144877. All other authors declare no competing interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION
_Nature Communications_ thanks the anonymous reviewer(s) for their contribution to the peer review of this work. PUBLISHER’S NOTE Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION REPORTING SUMMARY SOURCE DATA SOURCE DATA RIGHTS AND PERMISSIONS
OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or
format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or
other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in
the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the
copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Hennequart, M.,
Labuschagne, C.F., Tajan, M. _et al._ The impact of physiological metabolite levels on serine uptake, synthesis and utilization in cancer cells. _Nat Commun_ 12, 6176 (2021).
https://doi.org/10.1038/s41467-021-26395-5 Download citation * Received: 28 August 2020 * Accepted: 01 October 2021 * Published: 26 October 2021 * DOI:
https://doi.org/10.1038/s41467-021-26395-5 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative