The role of hybridisation in the origin and evolutionary persistence of vertebrate parthenogens: a case study of darevskia lizards
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Obligate parthenogenesis is found in only 0.1% of the vertebrate species, is thought to be relatively short lived and is typically of hybrid origin. However, neither the
evolutionary persistence of asexuality in vertebrates, nor the conditions that allow the generation of new parthenogenetic lineages are currently well understood. It has been proposed that
vertebrate parthenogenetic lineages arise from hybridisation between two divergent taxa within a specific range of phylogenetic distances (the ‘Balance Hypothesis’). Moreover,
parthenogenetic species often maintain a certain level of hybridisation with their closest sexual relatives, potentially generating new polyploid hybrid lineages. Here we address the role of
hybridisation in the origin and evolutionary lifespan of vertebrate parthenogens. We use a set of microsatellite markers to characterise the origins of parthenogens in the lizard genus
_Darevskia_, to study the distinctiveness of sexual and asexual taxa currently in sympatry, and to analyse the evolutionary consequences of interspecific hybridisation between asexual
females and sexual males. We find that parthenogens result from multiple past hybridisation events between species from specific lineages over a range of phylogenetic distances. This
suggests that the Balance Hypothesis needs to allow for lineage-specific effects, as envisaged in the Phylogenetic Constraint Hypothesis. Our results show recurrent backcrossing between
sexual and parthenogenic _Darevskia_ but neither gene flow nor formation of new asexual lineages. We suggest that, along with their demographic advantage, parthenogens gain additional
leverage to outcompete sexuals in nature when the retention of sexual reproductive machinery allows backcrossing with their sexual ancestors. You have full access to this article via your
institution. Download PDF SIMILAR CONTENT BEING VIEWED BY OTHERS NATURAL REPEATED BACKCROSSES LEAD TO TRIPLOIDY AND TETRAPLOIDY IN PARTHENOGENETIC BUTTERFLY LIZARDS (_LEIOLEPIS_: AGAMIDAE)
Article Open access 24 January 2025 PERVASIVE GENE FLOW DESPITE STRONG AND VARIED REPRODUCTIVE BARRIERS IN SWORDTAILS Article Open access 26 March 2025 MULTIPLE CONTACT ZONES AND KARYOTYPIC
EVOLUTION IN A NEOTROPICAL FROG SPECIES COMPLEX Article Open access 11 January 2024 INTRODUCTION Asexual reproduction is distributed across all major clades of the tree of life. Exclusively
asexual species are expected to be short lived (Otto 2009), typically have a ‘twiggy’ phylogenetic distribution (Bell 1982) and originate from sexual ancestors (Avise 2008). Many studies
have focused on the evolution of sex, trying to understand how such a costly reproductive mode is so successful in nature (Weismann 1889; Maynard Smith 1978; Otto 2009). This paradox of sex
has been tentatively explained by the superior potential for adaptation to changing environments of sexual species compared with asexual ones (McDonald et al. 2016; Luijckx et al. 2017).
Given the putative young age of (most) asexual species, we can deduce that sexual species constantly have the potential to give rise to new asexual lineages, but the predicted offset between
generation and extinction of asexual lineages has not often been studied empirically. The maintenance of asexual species (which constitute clusters of lineages of similar genotype and
phenotype; cf. Barraclough 2010) depends on the generation of asexual lineages from sexual progenitors and their loss, either through accumulation of deleterious mutations (Haigh 1978),
failure to adapt (Lively 2010) or neutral processes (Schwander and Crespi 2009; Janko 2014). Asexual invertebrates generally result from spontaneous thelytokous parthenogenesis (Bullini
1994). Parthenogenetic vertebrates are less common than invertebrate asexuals (Beukeboom and Vrijenhoek 1998), they are obligate, mostly recent (Beukeboom and Vrijenhoek 1998; Avise 2008)
and often include polyploids (Bullini 1994). Parthenogenetic vertebrates typically originate through hybridisation between two individuals of different taxa (e.g. Choleva et al. 2012; Lutes
et al. 2011; but see Sinclair et al. 2010), often resulting in sexual–asexual complexes including a network of species that recurrently hybridise (Danielyan et al. 2008), and eventually
allow for the origin of new parthenogenetic species (Taylor et al. 2015). Cases where gene flow occurs between proposed evolutionary units, or where there is a recurrent origin of new
asexual lineages from sexual parents, question the current definition and applicability of some species concepts (Coyne et al. 1988; Birky and Barraclough 2009). However, repeated
hybridisation events often underlie the origin of parthenogenetic vertebrate species (as in _Aspidoscelis_ sp. (Reeder et al. 2002), _Lepidodactylus lugubris_ (Trifonov et al. 2015) and
_Cobitis elongatoides-taenia_ (Choleva et al. 2012)). Since these events result in phenotypically and genetically similar lineages, they are consistent with the definition of asexual species
used here. Two general hypotheses have been put forward regarding the conditions required for interspecific hybridisation to give rise to parthenogenetic lineages: the Balance Hypothesis
(BH) (Wetherington et al. 1987; Moritz et al. 1989; an idea first discussed with reference to plants by Ernst 1918 (cited in Bartos et al. 2019)) and the Phylogenetic Constraint Hypothesis
(PCH) (Darevsky 1967; Murphy 2000). The BH proposes that parthenogenetic vertebrates arise by hybridisation between two sexual species divergent enough to disrupt meiosis in the hybrids, yet
not so divergent as to compromise hybrid viability or (parthenogenetic) fertility (Moritz et al. 1989; Kearney et al. 2009). Under the BH, pairwise distances between parental species, at
the inferred time of origin of the parthenogenetic hybrids, should fall within an interval that is constrained at both ends. Before that interval, viable and sexually fertile offspring are
formed, and after that window hybrids are either inviable or both sexually and parthenogenetically infertile. This window is likely to be much shorter than the time since the separation of
the parental lineages. Any pair of clades that has passed through this interval will have had the opportunity to generate parthenogenetic lineages. We consider this the core of the BH.
Moritz et al. (1989) further suggested that ploidy changes might alter the trade-off between incompatibility and disruption of meiosis, and so the window for the origin of parthenogenetic
lineages. We view this as an extension of the core BH. The PCH proposes that asexual lineages originate from hybridisation between the pairs of sexual species that, alone or in combination,
possess lineage-dependent genetic factors that allow them to interbreed and produce viable hybrids capable of reproducing parthenogenetically. One lineage may possess particular factors that
need to come from the maternal lineage, for example, associated with female meiosis or egg formation. In this case, the PCH also predicts that hybridisation events generating parthenogens
will be directional, with species from different clades contributing either the maternal or paternal ancestry (Murphy 2000). These two hypotheses are not mutually exclusive. Under the PCH,
hybridisation between very close or very distant lineages is unlikely to generate parthenogenesis. The BH does not exclude a higher probability of generating parthenogenetic lineages from
some combinations of parental species than others. Nevertheless, it is helpful to consider whether the origin of parthenogenetic lineages is mainly constrained by lineage-specific effects
(PCH) or by genetic distance (BH) because this might give insight into the phylogenetic distribution of asexual vertebrates and the conditions needed for their generation and persistence.
The model system used in this study, _Darevskia_ lizards, has a hybridisation-rich evolutionary history, and therefore is a suitable model to study the correlation between hybridisation and
parthenogenesis. All of its asexual lineages are reported to be of hybrid origin (Murphy 2000; Freitas et al. 2016), there is evidence for recurring mating between asexual females and sexual
males when in sympatry, generating polyploid backcrosses (Darevsky and Danielyan 1968; Danielyan et al. 2008) and for frequent interspecific hybridisation between sexual species (Darevsky
1967). Given that maternal and paternal ancestors of parthenogenetic _Darevskia_ are reported to be limited to a few sexual species, this model appears to fulfil the predictions of the PCH
(Darevsky 1967) although this observation alone does not exclude the BH. However, inferences of the origin of the different parthenogenetic species and the identification of the polyploids
have been based on limited evidence, and require genetic confirmation. Initial estimates of the phylogenetic relationships of sexual species (Murphy et al. 1996; Murphy 2000) suggest
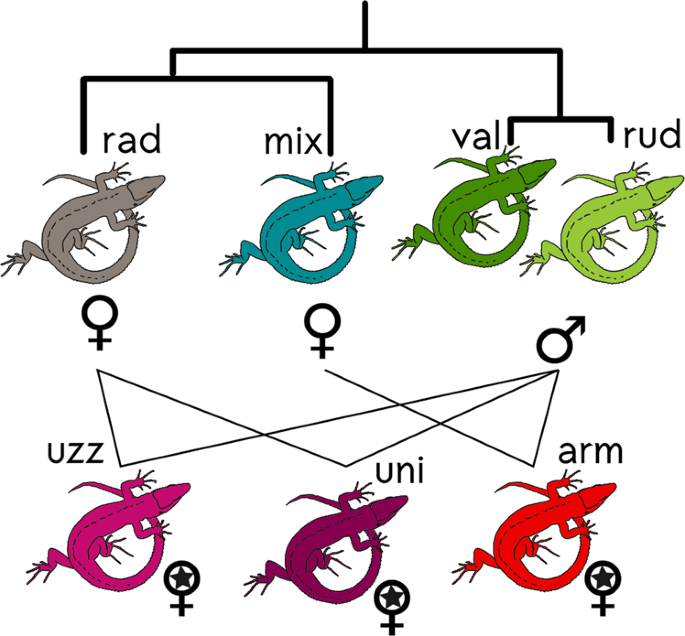
_Darevskia_ is divided into three main clades (Fu et al. 1997). Considering the asexual species and their putative parents (Fig. 1), _D. armeniaca_ is thought to have resulted from
hybridisation between _D. mixta_ and _D. valentini_ (Fu et al. 2000a, Moritz et al. 1992), while both _D. unisexualis_ and _D. uzzelli_ are thought to have resulted from hybridisation
between _D. raddei_ and _D. valentini_ (Fu et al. 2000b). The maternal species was always _D. raddei_ or _D. mixta_ (placed in the same unresolved terminal clade by Murphy 2000) and the
paternal species was _D. valentini_ or _D. portschinskii_ (the latter, and its descending asexual hybrid lineages, were not analysed in this study)_. Darevskia rudis_ is the third species
from the rudis clade, phylogenetically very close to _D. valentini_ and _D. portschinskii_, and there is evidence for between-species gene flow within this clade (Tarkhnishvili et al. 2013).
The relevance of _D. rudis_ to the origin of the parthenogens is still unclear. Hybridisation between parthenogenetic _Darevskia_ females and males of their sexual parental species has been
reported, based on morphology and karyology of a limited set of individuals (Danielyan et al. 2008). Reported triploid hybrids (3_n_ = 57) included both females and males, with unknown
fertility and varying levels of reproductive organ development (Danielyan et al. 2008). Since eggs of different ploidy may develop in the same oviduct, it is not clear if hybrids between
sexual males and asexual females are always polyploid or can also be diploid. The goal of this study was to address three questions related to the origin of parthenogenesis in vertebrates,
using _Darevskia_ as a model: (1) Is the origin of hybrid asexuality in _Darevskia_ more consistent with the BH or with the PCH? (2) Did multiple hybridisation events originate each
recognised parthenogenetic _Darevskia_ species? (3) Is there ongoing backcrossing with gene flow between the parthenogens and their sexual parents? To address these issues, we used
microsatellite markers and genotyped individuals from a wide range of localities from Armenia, Turkey, Georgia and Iran. We reassess the inferred parentage of three parthenogenetic
_Darevskia_: _D. unisexualis_, _D. uzzelli_ and _D. armeniaca_, comparing matrilineal (mtDNA) and nuclear (maternal + paternal) lineages of the putative parents and parthenogens. Current
sympatric localities were surveyed, and the evolutionary significance of hybrids is discussed. Our results indicate that the origin of parthenogenetic _Darevskia_ is more consistent with PCH
than with BH. We found no evidence that recurrent backcrossing between sexual and parthenogenetic _Darevskia_ has led to gene flow. Finally, our results suggest that, while they retain
sexual reproductive machinery, parthenogenetic vertebrates impose an additional cost on sympatric sexual populations by backcrossing with their sexual parents. MATERIAL AND METHODS SAMPLE
COLLECTION In this study, 378 _Darevskia_ individuals were analysed (Table S1, Fig. S1). Samples were collected between 2007 and 2011 in Armenia, Turkey, Georgia and Iran, aiming to include
representatives of each species’ distribution range. Seven species were included in the analyses, three parthenogenetic (_D. armeniaca_, _D. unisexualis_ and _D. uzzelli_) and four sexual
(_D. mixta_, _D. raddei_, _D. valentini_ and _D. rudis_). Individuals were provisionally identified in the field based on overall morphology, size, colour pattern and scutellation (Arakelyan
et al. 2011). Four sympatric localities in Armenia where sexual and parthenogenetic species coexist were sampled (Table S2). In Kuchak, previous studies have reported backcross individuals
between parthenogens and sexual species (Danielyan et al. 2008). Three species have been found: two parthenogenetic (_D. armeniaca_ and _D. unisexualis_) and one sexual (_D. valentini_), the
putative paternal species for both parthenogens present. Putative backcross individuals, _D. armeniaca_ × _D. valentini_ and _D. unisexualis_ × _D. valentini_, have been identified based on
morphology, and polyploidy of some individuals has been confirmed by karyology, identifying both triploids and tetraploids (Danielyan et al. 2008). In total, 150 individuals were sampled,
together with individual information (location, pictures, temperature) reported elsewhere (Carretero et al. 2018; Sillero et al. 2018). In Sotk, the sexual _D. valentini_ and the parthenogen
_D. armeniaca_ have been found in sympatry, and individuals morphologically similar to _D. armeniaca_ × _D. valentini_ backcrosses were tentatively identified based on large body size and
intermediate colouration. In the remaining two localities, Lchap and Lchashen, the sexual _D. raddei_ was found together with its hybrid descendant, the parthenogen _D. unisexualis_ (Lchap),
or with the sexual _D. valentini_ (Lchashen). GENOTYPING Genomic DNA was extracted from 30 mg of tail-tip tissue following standard high-salt protocols (Sambrook and Russell 2001). From a
total of 74 tested microsatellite loci developed previously for other lacertid lizards, 12 polymorphic markers were selected on the basis of reliable amplification and heterozygosity: D119,
C118, C113 (Remón et al. 2008), Pb55 (Pinho et al. 2004), Lv-4-72 (Boudjemadi et al. 1999), P011, P054 (Wellenreuther et al. 2009), Ph39, Ph124, Ph128, Ph170 (NCBI accession numbers:
KC869962, KC869964, KC869956, KC869961) and Du323, Du47, Du418 (Korchagin et al. 2007) (for more information see Table S3). PCR amplifications were carried out using the Multiplex PCR Kit
(QIAGEN) following the manufacturer’s instructions in a final 10 μl volume, including a negative control. Amplicons were separated by size on an ABI3130xl Genetic Analyzer. Allele sizes were
scored against the GeneScan500 LIZ Size Standard using GENEMAPPER 4.0 (Applied Biosystems) and manually checked twice, independently. To control against allelic dropout, which is expected
to be higher in polyploids due to the greater number of amplicons in each PCR reaction, 35–45% of genotypes per marker were repeated, including all the putative polyploids, in independent
PCR reactions (Table S3). For the repeated samples, loci were genotyped individually to confirm that the third (or fourth) allele scored was not an artefact of interaction between the
different primer pairs in the multiplex. To test for the presence of null alleles, genotyping errors and allelic dropout, we used Microchecker 2.2.3 (Van Oosterhout et al. 2004). These tests
were performed only on _D. valentini_ (excluding individuals from sympatric localities) because it was the species with most individuals, and the assumptions of this analysis are not
appropriate for parthenogenetic species or for mixed-species samples. Tests for Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD), and standard genetic diversity measures,
observed (HO) and expected (HE) heterozygosities, allele frequencies and allelic richness, were obtained using Cervus/Fstat v2.9.3.2 (Goudet 1995). The critical probability for each test was
adjusted with a sequential Bonferroni correction (Rice 1989). Only markers in HWE and without significant LD were used hereafter. SYMPATRIC LOCALITIES: PLOIDY DETERMINATION Individuals
found at the sympatric localities were analysed for their ploidy and parentage, testing the outcomes of hybridisation between parthenogenetic females and sexual males. Ploidy level was
identified as the maximum number of alleles found among the markers used (ploidy function in Polysat; (Clark and Jasieniuk 2011)). All individuals characterised as triploids had at least two
markers with three different alleles. Given the multiple ploidy levels in the sympatric localities (see “Results” section), traditional diversity measures and population genetic analyses
could not be applied to these datasets. Instead, we calculated inter-individual genetic distances using the Bruvo method (Bruvo et al. 2004) implemented in Polysat (detailed information in
SI). SYMPATRIC LOCALITIES: PRINCIPAL COORDINATE ANALYSIS Principal Coordinate Analyses (PCoA) were performed on the individuals from the sympatric localities, and then plotted against the
corresponding species from elsewhere in the distribution. Bruvo distances were used as a dissimilarity matrix, and sympatric localities were grouped in pairs according to their species
makeup (Table S2). Kuchak and Sotk constituted the first pair, both with _D. valentini_ (sexual paternal species) and parthenogenetic species, and Lchap and Lchashen the second, both with
_D. raddei_ (sexual maternal species) and others. CLUSTER ANALYSES Cluster analyses were performed on the dataset of all diploid individuals, using a hierarchical approach. First, we
determined the optimal number of clusters (_K_) on the dataset of all diploid individuals, excluding the diploids from the sympatric localities. _K_ was determined using the _find.clusters_
option in the ADEGENET package (Jombart et al. 2010) by comparing the different clustering solutions using a Bayesian Information Criterion (BIC). For similar BIC values, the optimal _K_
value was selected based on concordance between the clusters and described taxa. This value of _K_ was then compared with ∆_K_ and the rate of change of the log probability of the data
between successive _K_ values calculated from structure analyses on the same dataset (see below). Second, we visualised the relationships among sexual and parthenogenetic species using the
diploid individual dataset, now including the diploids from the sympatric localities. We performed an exploratory Discriminant Analysis of Principal Components (DAPC) also implemented in
ADEGENET (Jombart et al. 2010). We used the _K_ value calculated previously as the number of clusters to which individuals were assigned, and DAPC was used to ordinate individuals according
to axes that maximise cluster distances relative to variation within clusters. We also calculated the pairwise _F_ST values between the species (diploids from the sympatric localities were
separated into independent groups) in this same dataset with _pairwise.fst_ in the package hierfstat, version 0.04–22 (Goudet 1995). To avoid effects on _F_ST due to small sample size, the
pairwise _F_ST was estimated only when seven or more individuals were available per species and population. Third, we determined the numbers of (diploid) parthenogenetic lineages and hybrid
origins of the parthenogenetic diploid individuals (including diploid parthenogens from the sympatric localities) by conducting assignments using DAPC membership probability values for each
parthenogenetic species alone. To further confirm the number of hybrid origins per species, we calculated the Bruvo (Bruvo et al. 2004) pairwise distance distribution among individuals of
each parthenogenetic species. The Bruvo method was used here so that comparisons with polyploids were made possible. Lynch distances showed a similar arrangement of clusters and distances
between species and hybrids (Fig. S2, more information in the supplementary material). Hartigan’s dip test (implemented in R package diptest version 0.75–7:
https://CRAN.R-project.org/package=diptest) was used to assess departure from an unimodal distance distribution. Fourth, to test whether genetic structure was due to isolation by distance
(IBD) or the presence of distinct groups within taxa, IBD analysis was performed on the nominate sexual species _D. raddei, D. valentini_ and _D. mixta_ (including sympatric localities),
with the ADEGENET package in R (Jombart et al. 2010). Last, population structure and the ancestry of the parthenogens were investigated using the Bayesian multilocus clustering analysis
implemented in STRUCTURE v2.3.4 (Pritchard et al. 2000; Hubisz et al. 2009) (for run details see SI). The sexual species (diploid individuals from sympatric localities included) were used as
‘learning samples’ (PopFlag = 1) (Murgia et al. 2006) to define the cluster membership when, subsequently, diploid parthenogenetic individuals were included in the dataset. Cluster
membership was assigned according to the optimal number of clusters calculated previously (_K_), and excluding the clusters of parthenogens (since only sexual species were used as ‘learning
samples’). Given their hybrid origin, parthenogens are expected to have half of their ancestry from the maternal species and the other half from the paternal species, with little variation
due to their clonal reproduction. The ancestry of the parthenogenetic individuals from sympatric and allopatric localities (and two diploid individuals of uncertain status) was determined
(PopFlag = 0) (Pritchard et al. 2000). Some level of misclassification was allowed with the MIGRPRIOR set at 0.01. RESULTS GENOTYPING: GENERAL OVERVIEW All analyses were performed with 12
markers in total, nine of which were discriminating for the polyploids. The remaining three markers never presented more than two alleles in any of the inferred polyploids. Although
surprising, this is compatible with the alleles present in the putative parental populations for these three loci (Table S4). Low levels of uncertainty were found (<1% disagreement
between repeats) and uncertain genotypes were eliminated from further analyses. Polyploid individuals were found only in sympatric localities and classified as hybrids between parthenogens
and sexuals (hereafter ‘PS hybrids’). Thirteen individuals presented three alleles only for one marker (Table S1). These individuals may have been triploids with unusually low
heterozygosity, or there may have been genotyping errors but it is likely they were aneuploid, since aneuploidy has been observed previously in this genus (Kupriyanova 2009). Given the
purpose of this study, we followed a conservative approach and kept these individuals in downstream analyses as diploids, with the specific markers assigned as missing data. Ploidy
determination and the exploratory DAPC identified some putative misidentifications (details in SI). These individuals were kept in the analysis and reclassified according to their genetic
determination, followed by revised DAPC analyses (Fig. S3). SEXUAL AND PARTHENOGENETIC SPECIES STRUCTURE The optimal number of clusters was selected after comparison of the BIC values from
DAPC with the best _K_ values in STRUCTURE. When setting clusters for the sexual populations only, _K_ = 7 resulted in the most consistent groups in both the DAPC and STRUCTURE analyses.
These clusters corresponded to the recognised species, but also distinguished groups within species. In _D. valentini_ and _D. raddei_ this was probably due to spatial structure, reflected
in significant isolation by distance (Figs. S4 and S5). _Darevskia mixta_ consistently fell into two well differentiated and geographically separated groups (_D. mixta-1_ and _D. mixta-2_),
with an _F_ST distance (_F_ST = 0.25, Table 1) similar to, or even higher than, other recognised species pairs. These subgroups within species were kept separate since they may be relevant
to determining the parentage of parthenogens. All sexual species showed heterozygote deficiencies (Fig. 2), most likely due to merging of geographically variable populations. When including
the three parthenogenetic species, they were consistently divided into four clusters in DAPC (two clusters for _D. armeniaca_). Therefore, a _K_ value of 11 was used in the DAPC of the total
diploid dataset (sexual + parthenogenetic, including diploids of the sympatric localities). Parthenogens (_D. unisexualis_, _D. uzzelli_ and _D. armeniaca_) had higher observed than
expected heterozygosity, as predicted due to their hybrid origin (Fig. 2). All parthenogenetic species had private alleles, absent from the sexual taxa and sometimes in high frequencies
(e.g. allele 258 represents 53% of the diversity of the C113 marker in _D. uzzelli;_ Table S5). ASEXUAL PARENTAGE INFERENCE STRUCTURE analysis following training on the sexual species (_K_
_=_ 7) clearly showed a mixed ancestry for each of the parthenogenetic genomes (Fig. 3, Table S6). _Darevskia armeniaca_ shared approximately half of its genome with _D. mixta-1_ and the
other half was divided between _D. valentini_ and _D. rudis_ (predominantly from one cluster within each of these sexual species). Similarly, _D. uzzelli_ shared half of its genome with _D.
raddei_ and the other half with a mix of _D. valentini_ and _D. rudis_ clusters. The _D. unisexualis_ genome was also shared half with _D. raddei_ and the other half with the _D. valentini_
and _D. rudis_ clusters, in a similar but not identical composition to _D. uzzelli_. Mitochondrial DNA sequences confirmed previous maternal parent assignments (Fig. S6; see SI for methods).
Across all three parthenogens, the contribution from the putative maternal species was closer to 50% (0.36–0.58) than the contribution from the proposed paternal species (0.10–0.45). This
is reflected in lower _F_ST between each parthenogen and its maternal parent than its putative paternal ancestor (Table 1). ORIGIN OF PARTHENOGENETIC HYBRIDS: NUMBER OF HYBRIDISATION EVENTS
Analyses of the distributions of Bruvo distances were performed to assess the number of origins of each parthenogenetic species. A parthenogenetic species with a single origin is expected to
display a unimodal distribution whose variance increases with lineage age and might become fragmented due to spatial structure and extinction of lineages. On the other hand, more than one
origin might generate a multimodal distribution dependent on sampling of genotypes from the parental populations. The _D. armeniaca_ distance distribution (Fig. 4, inset A) not only extended
over a wider range than the remaining parthenogens, but also was bimodal (with the first peak more marked than the second; Hartigans’ dip test D = 0.0657, _p_-value < 2.2e−16). In the
DAPC analysis (Fig. S7), Kuchak individuals and those from the remaining distribution were separated into two groups, a separation possibly accentuated by the overrepresentation of Kuchak
individuals in the dataset. Nevertheless, the presence of a higher number of private alleles (Table S5) in the population from Kuchak, in comparison to the remaining distribution, suggests
that this population has had more time to accumulate new alleles than the rest of the distribution. These observations are consistent with _D. armeniaca_ being older than the other
parthenogens and possibly having more than one origin for extant clones. Bruvo distances for _D. uzzelli_ were distributed over a range of distances similar to _D. armeniaca_, there was a
significant departure from unimodality (Hartigans’ dip test D = 0.0528, _p_-value < 2.2e−16) and a significant bimodality. Despite the wide interval of distances, _D. uzzelli_ was the
parthenogen with the lowest number of private alleles. Finally, _D. unisexualis_ had one peak in the Bruvo distance distribution, and the narrowest range of distances among the three
parthenogens analysed. Hartigans’ dip test indicated departure from unimodality (D = 0.03065, _p_-value = 0.002), but the distribution was unimodal at low resolution and ragged at higher
resolution; it also indicated multimodality with at least four peaks. A bimodal pattern was recovered in the DAPC analysis. _Darevskia unisexualis_ from Kuchak also had private alleles
(Table S5) but, in this species, more private alleles were found in the remaining distribution (unlike _D. armeniaca_). SYMPATRIC LOCALITIES Polyploid individuals were found in three of the
sympatric localities, Kuchak, Sotk and Lchashen. Following ploidy assignment in Kuchak, 17% (27/160) were polyploids and interpreted as parthenogenetic × sexual (PS) hybrids: 3% (5/160) _D.
armeniaca_ × _D. valentini_ and 14% (22/160) _D. unisexualis_ × _D. valentini_. Diploid individuals from Kuchak belonged to the parthenogens _D_. u_nisexualis_ and _D. armeniaca_, and the
sexual _D. valentini_. One tetraploid was found in Kuchak and another in Sotk, both PS hybrids _D. armeniaca_ × _D. valentini_ (IDs 12176 and 9910, respectively: Table S1). In Sotk, 11
individuals were _D. armeniaca_ × _D. valentini_ triploids and five _D. valentini_ sexual diploids. In the PCoA analysis of Kuchak and Sotk (Fig. 5a), a total of 277 individuals were
included: 160 from Kuchak and 25 from Sotk, the remainder from the general distribution range (Table S1). _Darevskia valentini_ formed a separate cluster from each of the two parthenogens,
_D. armeniaca_ and _D. unisexualis_, and the three clusters were approximately equidistant. Diploid individuals from Kuchak and Sotk mostly overlapped with the distribution of genotypes from
elsewhere in the corresponding species’ ranges, with some exceptions (specifically among _D. armeniaca_ and _D. valentini_ from Kuchak). As expected, genotypic variation was greater in the
sexual species than the parthenogens. Triploid _D. armeniaca_ × _D. valentini_ PS hybrids from Kuchak and Sotk fell between their proposed maternal (the parthenogen _D. armeniaca_) and
paternal species (the sexual _D. valentini_). As expected, these clusters were closer to the parthenogenetic parent, which contributed two out of three alleles. Triploid _D. unisexualis_ ×
_D. valentini_ PS hybrids from Kuchak behaved similarly, clustering between their proposed maternal (the parthenogen _D. unisexualis_) and paternal species (the sexual _D. valentini_). Both
tetraploid individuals fell among the remaining PS hybrids of the same cross (Fig. 5a). Bruvo distances among the PS hybrids were distributed over a narrower interval for _D. unisexualis_ ×
_D. valentini_ than for _D. armeniaca_ × _D. valentini_ hybrids, even though there were many more _D. unisexualis_ × _D. valentini_ comparisons (Fig. 4, bottom panels). Average pairwise
distances were generally larger for the polyploids than for the diploid parthenogens (Fig. 4, top panel), and a pairwise distance of zero was never found among the polyploids. If the
triploid PS hybrids originate via recurrent backcrossing each generation between the asexual females and the sexual males, they are expected to carry only alleles that are shared with their
parthenogenetic maternal species (_D. armeniaca_ or _D. unisexualis_, which are also of hybrid origin) or with the paternal species (_D. valentini_). As predicted, we found that PS hybrids
carried one allele unique to _D. valentini_ and the other (one or) two either unique to the parthenogen (_D. unisexualis_ or _D. armeniaca)_ or shared by the parthenogen and _D. valentini_.
The allelic combination for each marker was normally such that the source of each allele could be easily determined. Only two _D. unisexualis_ × _D. valentini_ triploid PS hybrids (IDs 12182
and 12574) had unique alleles (Table S5), one allele each that was absent from all other species in this study. In the PCoA with the Lchap and Lchashen (Fig. 5, panel b), only 19
individuals in total were available (Table S1). All _D. raddei_ individuals from these localities clustered with _D. raddei_ from elsewhere in its range, and separately from the _D.
unisexualis_ cluster found in Lchap. _D. valentini_ individuals fell close to, but not within the cluster formed by _D. valentini_ individuals from elsewhere, and unexpectedly not very close
together. The PS hybrid _D. armeniaca_ × _D. valentini_ found in Lchashen (red triangle on the Fig. 5, panel b) fell within the cluster formed by the PS hybrids _D. armeniaca_ × _D.
valentini_ from Sotk when analysed together (results not shown). Diploids from the sympatric localities were included in the structure analysis and the genomic contributions of parents to
these individuals were concordant with the same species from other non-sympatric localities. The proportional contributions from the putative parents were slightly different between _D.
armeniaca_ from Kuchak and from elsewhere (Table S6), consistent with weak bimodality in the Bruvo pairwise genetic distance distribution (Fig. 4, top panel) and DAPC. The contributions
varied little among Kuchak individuals, indicating the presence of one abundant clone (and generating the peak at low Bruvo distance). DISCUSSION Our study focused on _Darevskia_ rock
lizards as a model for the origin and dynamics of parthenogenesis in vertebrates. Our results support previous parentage assignments for the parthenogens. Only two lineages act as parents,
one consistently as the maternal parent. This pattern is more compatible with the PCH than with the core BH. We present evidence for multiple origins of each parthenogenetic species. Our
results also show ongoing hybridisation between parthenogenetic females and sexual males, but no gene flow was detected among the interbreeding species. The sexual (and diploid) _Darevskia_
lizards analysed here were divided into genetically distinct groups, concordant with the initial assignment based on phenotype. _Darevskia mixta_ individuals were consistently divided into
two genetic groups, one with samples from Georgia (_D. mixta-1_) and the other with samples from Turkey (_D. mixta-2_). Pairwise _F_ST between these groups was 0.25, similar to the distance
between other sexual species pairs, suggesting species-level differentiation. With structure analysis, we reassessed the parentage inferences made by Darevsky (1967), Fu et al. (2000a,
2000b) and Murphy (2000) and confirmed the hybrid genetic profile of all diploid parthenogenetic species studied. This analysis was performed with the allelic information from 12
microsatellite loci only, so it is likely to have some uncertainty. However, lack of LD among the loci suggests that they are not closely linked and so provide a meaningful estimate of the
proportional contributions of parental genomes. We found that the maternal contribution (confirmed by mtDNA analysis, Fig. S6) for each parthenogen (either _D. raddei_ or _D. mixta_-1)
accounted for ~50% of the nuclear genome of the parthenogenetic species, as expected. On the other hand, the paternal contribution was less clear (Fig. 3). For each of the three
parthenogens, there appeared to be a mix of contributions from the sexual species _D. valentini_ and _D. rudis. Darevskia valentini_ was the previously proposed paternal species in all these
cases (Darevsky 1967). On the basis of five microsatellite markers, Tarkhnishvili et al. (2017) claimed to provide evidence for the origin of some _Darevskia_ parthenogenetic lineages (e.g.
_D. armeniaca_) from backcross hybridisation between one parthenogenetic female and a male of a sexual species (Tarkhnishvili et al. 2017). However, that would not explain the 50% maternal
genomic ancestry found in all parthenogenetic individuals analysed here, since it would imply triploidy and a contribution of ~33%. More parsimonious explanations are available: a prior
hybridisation between two sexual individuals of different species, in this case _D. rudis_ and _D. valentini_ (for which there is some evidence; Tarkhnishvili et al. 2013), could have
generated a sexual hybrid with a mixed genome that then hybridised with a female of a different sexual species (either _D. raddei_ or _D. mixta_-1), leading to the origin of an asexual
hybrid lineage with contributions from three parental taxa. For this to have happened, the three different species would have to have been in sympatry, a scenario facilitated by the frequent
secondary contacts _Darevskia_ taxa experienced during the Pleistocene ice ages (Freitas et al. 2016). Moreover, widespread evidence for gene flow among sexual taxa (Freitas et al.
unpublished), including among species of the rudis clade (_D. valentini_, _D. rudis_ and _D. portschinskii_) (Tarkhnishvili et al. 2013), further supports this suggestion. Ancient
hybridisation appears to fuel speciation events in other taxa as well (e.g. Meier et al. 2017). We cannot rule out contributions from unsampled populations, but we conclude that prior
hybridisation is the most likely explanation for our findings. (1) IS THE ORIGIN OF HYBRID ASEXUALITY IN _DAREVSKIA_ MORE CONSISTENT WITH THE BALANCE HYPOTHESIS OR WITH THE PHYLOGENETIC
CONSTRAINT HYPOTHESIS? Under the BH (Moritz et al. 1989), parental species pairwise distances, at the inferred time of origin of the parthenogenetic hybrids, should fall within a constrained
interval (Kearney et al. 2009). Any species pair that has passed through that interval could have generated parthenogenetic hybrids but it is likely that only a small proportion of them
will give rise to lineages that are now extant. In _Darevskia_, we used _F_ST pairwise distances calculated from the 12 microsatellite markers. Given the small number of markers used and the
potential for homoplasy, distances may be inaccurate, particularly under-estimating longer distances. But, given the young age of the parthenogens (Freitas et al. 2016), current parental
distances are not much greater than distances when hybridisation gave rise to them. From these data we cannot assess constraints on the divergence interval during which parthenogens can be
formed. However, there are sexual species pairs with estimated distances within the same interval as the sexual parentals and with geographical range overlap (e.g. _D. mixta-1_/_D. rudis_ in
Western Caucasus). No parthenogenetic hybrid between these species has been reported. This conclusion also holds for distances based on mtDNA sequences (Table 1). The 23 sexual species of
_Darevskia_ not included in this study produced no known parthenogenetic hybrid species despite extensive survey effort with this group (Murphy 2000). This pattern is unexpected if
phylogenetic distance is the sole determinant of the potential for sexual species to give rise to parthenogenetic hybrids. On the other hand, it is the expected pattern under the PCH. More
precise distance estimates, based on more sequence data, and for a larger sample of species, will be needed to investigate this pattern further. We also confirm that the directionality of
the hybridisation events was consistent, _D. mixta_-1 and _D. raddei_ always acting as maternal species. This pattern is not expected under the core BH. While it is not a necessary feature
of the PCH, it can arise because the predisposition of particular lineages to generate parthenogenetic hybrids may be specific to either the maternal or paternal role. If such a
predisposition is maintained in the parthenogens, it may help to explain their observed ability to produce viable offspring with the sexual males of their paternal species. Despite the
viability of the offspring between parthenogenetic females and sexual males, the resulting hybrids are polyploid and apparently infertile, incapable of generating a stable parthenogenetic
lineage. The infertility of the polyploid hybrids could be explained under the extended BH, which proposes that the addition of a second haploid genome from one of the parental species can
disrupt the balance between meiotic disruption and incompatibility that allows the diploid hybrids to reproduce parthenogenetically. However, this hypothesis also supports the alternative
possibility (Moritz et al. 1989; Fig. 8), that the addition of a third genome can increase the fecundity and viability of the asexual hybrids (allowing a polyploid lineage to be generated).
Similarly, change in ploidy could have either effect on the lineage-specific factors underlying parthenogenesis postulated by the PCH. Thus, the infertility of the _Darevskia_ polyploids
does not provide evidence to distinguish between the BH and PCH. The BH remains the most widely accepted explanation for the origin of vertebrate parthenogenesis (Avise 2008). However, in
addition to _Darevskia_, other hybrid parthenogenetic vertebrates question whether the core BH is widely applicable. The hybridisation events that gave rise to the parthenogenetic
_Leiolepis_ (butterfly lizard) are directional and constrained to two clades within the genus (Grismer et al. 2014). In _Aspidoscelis_, the parthenogenetic hybrids always have a member of
the Sexlineatus clade as at least one of the parental species (Darevsky et al. 1985; Reeder et al. 2002). Still in this same genus, the parental pairs of the parthenogenetic hybrids are
composed both of species within the same phylogenetic clade (Lemniscatus and Sexlineatus groups), and species from different phylogenetic clades (Cozumela and Tesselata groups), and
consequently the distances among the parental pairs can be highly variable (Reeder et al. 2002; and see Janko et al. 2018 for a review showing variable distances between parental pairs of
asexual hybrids in Teleostei) suggesting little constraint on the divergence interval. Our results and other studies suggest that the distance between parental species is not as critical as
the specific characteristics of the sexual ancestors, and in some cases also the directionality of the hybridisation events. Some constraints on the interval of divergence, as proposed under
the BH, must apply. However, lineage-specific factors also appear to be critical to explaining the origin of parthenogenetic hybrid lineages. (2) DID MULTIPLE HYBRIDISATION EVENTS GIVE RISE
TO EACH PARTHENOGENETIC _DAREVSKIA_ SPECIES? The parthenogenetic _Darevskia_ species were initially described based on their morphological traits (Darevsky 1967; Arakelyan et al. 2011). The
number of hybridisation events giving rise to _Darevskia_ extant lineages of parthenogenetic species has been inferred to be one (_Darevskia rostombekowi_, Ryskov et al. 2017) or more than
one (_D. dahli_, Vergun et al. 2014). Our data show significant departures from unimodal distance distributions among genotypes for all three parthenogens. Given that these species are all
young, this is more likely to be a signal of multiple origins than diversification and fragmentation of a single lineage over time. However, more data and demographic modelling will be
needed to distinguish these possibilities. The three species analysed here behave differently in other respects. In _D. armeniaca_, which has a wide geographic range, the pairwise Bruvo
distance interval was wide and several private alleles were observed (most of them in Kuchak). _Darevskia unisexualis_ also has a wide range but its distance interval was narrower and
private alleles were not localised. Despite its young age (originated ~100 kya), _D. unisexualis_ is distributed over a relatively wide area (Freitas et al 2016). This rapid colonisation,
perhaps combined with competitive interactions between the _Darevskia_ parthenogens and their sexual relatives (Tarkhnishvili et al. 2010), can lead to local extinctions or expansions of
clones and so spatial heterogeneity in the parthenogenetic species. The multimodality recovered in the Hartigan’s test could be due to this type of clonal structure. _Darevskia uzzelli_ has
a narrow distribution range but the Bruvo distance interval was as wide as in _D. armeniaca_, despite it having few private alleles. _Darevskia armeniaca_ may have resulted from older
hybridisation events than the other two species. However, a single origin of extant clones followed by mutation accumulation in a patchy spatial distribution could also explain the pattern
observed. Evidence in other species (such as sexual trait decay, reviewed in van der Kooi and Schwander 2014) suggests that if the lower number of PS hybrids between _D. armeniaca_ and _D.
valentini_ in Kuchak is a result of sexual trait decay, the Kuchak population might be older than the Sotk population. In localities where the sexual parent species are currently in
sympatry, as in Lchashen, no evidence of new parthenogenetic lineages (or any parthenogenetic diploid individual) was found. Thus, there is no evidence that new parthenogenetic hybrids are
being generated at present, although a low rate of origin cannot be excluded. The origin of parthenogenetic _Darevskia_ might have been associated with secondary contacts promoted by the
last glaciations, as suggested on the basis of phylogeographic patterns and ecological niche models (Freitas et al. 2016). (3) IS THERE ONGOING BACKCROSSING WITH GENE FLOW BETWEEN THE
ASEXUALS AND THEIR SEXUAL PARENTALS? When hybridising with their sexual ancestors, PS hybrids frequently form new polyploid asexual lineages with hybridisation-induced parthenogenesis (e.g.
whiptail lizards _Aspidoscelis_ sp.: Cole et al. 2014; Taylor et al. 2015; Cole et al. 2017, butterfly lizards _Leiolepis_ sp.: Grismer et al. 2014; and the salamander _Ambystoma_ sp.: Bi
and Bogart 2010). However, it is also possible that triploid hybrids are formed in each generation and never establish polyploid lineages. Our results are more consistent with recurrent
backcrossing between sexual and parthenogenetic _Darevskia_ than with the existence of polyploid asexual lineages. Firstly, the pairwise distance distributions of the backcross polyploid
hybrids (PS hybrids) match the pairwise distance distributions of simulated genotypes produced by recurrent backcrossing (Fig. S8). Secondly, we did not recover any PS hybrid pair of
individuals with the same combination of genotypes for all markers, which we would find if the PS hybrids reproduced clonally. The rare private alleles carried by the PS hybrids are likely
due to limited sampling, or possibly genotyping error. The reproductive organs of PS hybrids from the populations studied here have been found to be undeveloped (Danielyan et al. 2008).
However, some of the triploids analysed here did show apparently normal reproductive organs or evidence of having already laid eggs (such as triploid female ID 10034 from Sotk). Triploid
hybrid males with apparently normal reproductive systems have already been described in this locality (Darevsky et al. 1985). Thus, the tetraploid individuals we found are likely to be the
result of a cross between a triploid and a diploid individual (either sexual _D. valentini_ or parthenogen _D. unisexualis_ and _D. armeniaca_). Both observations suggest that some triploids
could be partially fertile, although offspring viability is unknown. Traits required for sexual reproduction will be lost over time in asexual lineages (reviewed in van der Kooi and
Schwander 2014), with some reported cases of rapid loss (e.g. Lehmann et al. 2011). In _Darevskia_, the parthenogenetic lineages are young (around 100 kyr old; Freitas et al. 2016) and the
generation of triploid PS hybrids shows that they retain the sexual machinery necessary to mate and produce zygotes with a paternal contribution. However, these functions are expected to
decay with lineage age (van der Kooi and Schwander 2014). In Kuchak, we found a high ratio of _D. unisexualis_ × _D. valentini_ PS hybrids relative to _D. armeniaca_ × _D. valentini_,
concordant with previous findings (4:1 ratio in Danielyan et al. 2008). The reproductive pressure inferred from the intensity of copulation marks of _D. valentini_ males on each
parthenogenetic species present in Kuchak is the same (Carretero et al. 2018). Therefore, one possible explanation for the high proportion of _D. armeniaca_ × _D. valentini_ PS hybrids found
in Sotk, and low proportion of this cross found in Kuchak in relation to the other PS hybrid, is that the _D. armeniaca_ lineage present in Kuchak is older and has lost part of the sexual
reproduction machinery. This is concordant with the multimodal Bruvo distance distribution for _D. armeniaca_, suggesting that it had more than one origin through hybridisation events
separated in time and/or space. EVOLUTIONARY CONSEQUENCES OF HYBRIDISATION AND POLYPLOIDY IN _DAREVSKIA_ Asexuality in vertebrates can be a stage in the speciation continuum, providing an
effective barrier to gene flow when other forms of pre- and post-zygotic reproductive isolation are absent (Janko et al. 2018). We identified many polyploid hybrids (17%) in sympatric
locations of parthenogenetic species with their paternal sexual ancestors. Nevertheless, sexual and asexual species in the sympatric localities maintain their distinctiveness, suggesting
there is an effective barrier to gene flow. Parthenogenetic _Darevskia_ species may outcompete their sexual ancestors (Tarkhnishvili et al. 2010), or be sole occupants of habitats suitable
for their parentals (Freitas et al. 2016). When in contact, hybridisation between parthenogens and sexual males could reduce the number of sexual offspring in each generation. In this way,
sexual populations could experience an additional load, especially when in low abundance relative to parthenogens. This demographic vortex might lead sexual species towards local extinction.
Together with the two-fold reproductive output advantage of parthenogens compared with sexuals, hybridisation between the two forms could contribute to the out-performance by some
parthenogenetic species of their sexual ancestors. In summary, our study _of Darevskia_ as a model of vertebrate parthenogenesis questions whether the BH, suggested as a general theory for
the origin of hybrid asexuality, is sufficient to explain patterns in the origin of hybrid asexuality. Parthenogenesis in vertebrates is rare and generally originates from hybridisation
between specific species pairs with highly variable phylogenetic distances but often consistent maternal parentage. This fits the PCH that the parental species must hold some
lineage-dependent characteristics that allow them, when hybridising, to generate offspring capable of reproducing asexually. Identifying the constraining factors underlying the origin of
parthenogenetic vertebrates gives us a better chance of understanding how these hybrids use the sexual reproduction machinery to reproduce asexually, escaping the limitations that sex might
carry. DATA ARCHIVING Microsatellite genotypes: Dryad https://doi.org/10.5061/dryad.nk410t9 mtDNA sequences: GenBank MN210937 – MN211144, MN211145. CHANGE HISTORY * _ 23 SEPTEMBER 2019 An
amendment to this paper has been published and can be accessed via a link at the top of the paper. _ REFERENCES * Arakelyan M, Danielyan F, Corti C, Sindaco R, Levinton AE (2011) _The
herpetofauna of Armenia and Nagorno-Karabakh_. Society for the Study of Amphibians and Reptiles: San Francisco Google Scholar * Avise JC (2008) Clonality: the genetics, ecology, and
evolution of sexual abstinence in vertebrate animals. Oxford University Press, Oxford: New York, NY * Barraclough TG (2010) Evolving entities: towards a unified framework for understanding
diversity at the species and higher levels. _Philos Trans R Soc B_ 365:1801–1813 Article Google Scholar * Bartoš O, Röslein J, Kotusz J, Pačes J, Pekárik L, Petrtýl M et al. (2019) The
legacy of sexual ancestors in phenotypic variability, gene expression and homoeolog regulation of asexual hybrids and polyploids. _Mol Biol Evol_. https://doi.org/10.1093/molbev/msz114
Article PubMed PubMed Central Google Scholar * Bell G (1982) _The masterpiece of nature: the evolution and genetics of sexuality._ University of California Press: Berkeley Google Scholar
* Beukeboom LW, Vrijenhoek RC (1998) Evolutionary genetics and ecology of sperm-dependent parthenogenesis. _J Evol Biol_ 11:755–782 Article Google Scholar * Bi K, Bogart JP (2010) Time
and time again: unisexual salamanders (genus _Ambystoma_) are the oldest unisexual vertebrates. _BMC Evol Biol_ 10:238 Article PubMed PubMed Central CAS Google Scholar * Birky CW,
Barraclough TG (2009) Asexual Speciation. In: Schön I, Martens K, Dijk P (eds) _Lost sex: the evolutionary biology of parthenogenesis._ Springer Netherlands: Dordrecht, p 201–216 Chapter
Google Scholar * Boudjemadi K, Martin O, Simon J-C, Estoup A (1999) Development and cross-species comparison of microsatellite markers in two lizard species, _Lacerta vivipara_ and
_Podarcis muralis_. _Mol Ecol Notes_ 8:513–525 * Bruvo R, Michiels NK, D’souza TG, Schulenburg H (2004) A simple method for the calculation of microsatellite genotype distances irrespective
of ploidy level. _Mol Ecol_ 13:2101–2106 Article CAS PubMed Google Scholar * Bullini L (1994) Origin and evolution of animal hybrid species. _Trends Ecol Evol_ 9:422–426 Article CAS
Google Scholar * Carretero MA, García-Muñoz E, Argaña E, Freitas S, Corti C, Arakelyan M et al. (2018) Parthenogenetic females mate if males are around. An analysis of copulation marks in a
_Darevskia_ mixed community. _J Nat Hist_ 52:405–413 * Choleva L, Janko K, De Gelas K, Bohlen J, Šlechtová V, Rábová M et al. (2012) Synthesis of clonality and polyploidy in vertebrate
animals by hybridization between two sexual species: synthesis of clonality and polyploidy in vertebrate animals. _Evolution_ 66:2191–2203 Article PubMed Google Scholar * Clark LV,
Jasieniuk M (2011) polysat: an R package for polyploid microsatellite analysis. _Mol Ecol Resour_ 11:562–566 Article PubMed Google Scholar * Cole CJ, Taylor HL, Baumann DP, Baumann P
(2014) Neaves’ whiptail lizard: the first known tetraploid parthenogenetic tetrapod (Reptilia: Squamata: Teiidae). _Breviora_ 539:1–20 Article Google Scholar * Cole CJ, Taylor HL, Neaves
WB, Baumann DP, Newton A, Schnittker R et al. (2017) The second known tetraploid species of parthenogenetic tetrapod (Reptilia: Squamata: Teiidae): description, reproduction, comparisons
with ancestral taxa, and origins of multiple clones. _Bull Mus Comp Zool_ 161:285–322 Article Google Scholar * Coyne JA, Orr HA, Futuyma DJ (1988) Do we need a new species concept? _Syst
Zool_ 37:190–200 Article Google Scholar * Danielyan F, Arakelyan M, Stepanyan I (2008) Hybrids of _Darevskia valentini_, _D. armeniaca_ and _D. unisexualis_ from a sympatric population in
Armenia. _Amphib-Reptil_ 29:487–504 * Darevsky IS, Kupriyanova LA, Uzzell T (1985) Parthenogenesis in reptiles. In: Gans C, Billett F (eds) _Biology of the reptilia_. Wiley: New York, NY,
vol. 15, 411–526 Google Scholar * Darevsky I (1967) Rock lizards of the Caucasus: systematics, ecology and phylogenesis of the polymorphic groups of Caucasian rock lizards of the subgenus
Archaeolacerta. Nauka, Leningrad, Doctoral Thesis Google Scholar * Darevsky IS, Danielyan F (1968) Diploid and triploid progeny arising from natural mating of parthenogenetic _Lacerta
armeniaca_ and _L. unisexualis_ with Bisexual _L. saxicola valentini_. _J Herpetol_ 2:65 Article Google Scholar * Ernst A (1918) _Bastardierung als Ursache der Apogamie im Pflanzenreich.
Eine Hypothese zur experimentellen Vererbungs- und Abstammungslehre_. Nabu Press: Rudolstadt * Freitas S, Rocha S, Campos J, Ahmadzadeh F, Corti C, Sillero N et al. (2016) Parthenogenesis
through the ice ages: a biogeographic analysis of Caucasian rock lizards (genus _Darevskia_). _Mol Phylogenet Evol_ 102:117–127 Article PubMed Google Scholar * Fu J, MacCulloch RD, Murphy
RW, Darevsky IS, Tuniyev BS (2000a) Allozyme variation patterns and multiple hybridization origins: clonal variation among four sibling parthenogenetic Caucasian rock lizards. _Genetica_
108:107–112 * Fu J, Murphy RW, Darevsky IS (1997) Toward the phylogeny of Caucasian rock lizards: implications from mitochondrial DNA gene sequences (Reptilia: Lacertidae). _Zool J Linn Soc_
120:463–477 Article Google Scholar * Fu J, Murphy RW, Darevsky IS, McEachran JD (2000b) Divergence of the cytochrome _b_ gene in the _Lacerta raddei_ complex and its parthenogenetic
daughter species: evidence for recent multiple origins. _Copeia_ 2:432–440 Article Google Scholar * Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-Statistics. _J
Hered_ 86:485–486 Article Google Scholar * Grismer JL, Bauer AM, Grismer LL, Thirakhupt K, Aowphol A, Oaks JR et al. (2014) Multiple origins of parthenogenesis, and a revised species
phylogeny for the Southeast Asian butterfly lizards, _Leiolepis_. _Biol J Linn Soc_ 113:1080–1093 Article Google Scholar * Haigh J (1978) The accumulation of deleterious genes in a
population—Muller’s Ratchet. _Theor Popul Biol_ 14:251–267 Article CAS PubMed Google Scholar * Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure
with the assistance of sample group information. _Mol Ecol Resour_ 9:1322–1332 Article PubMed PubMed Central Google Scholar * Janko K (2014) Let us not be unfair to asexuals: their
ephemerality may be explained by neutral models without invoking any evolutionary constraints of asexuality. _Evolution_ 68:569–576 Article PubMed Google Scholar * Janko K, Pačes J,
Wilkinson-Herbots H, Costa RJ, Röslein J, Drozd P et al. (2018) Hybrid asexuality as a primary postzygotic barrier between nascent species: on the interconnection between asexuality,
hybridization and speciation. _Mol Ecol_ 1:248–263 Article PubMed PubMed Central Google Scholar * Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components:
a new method for the analysis of genetically structured populations. _BMC Genet_ 11:94 Article PubMed PubMed Central Google Scholar * Kearney M, Fujita MK, Ridenour J (2009) Lost sex in
the reptiles: constraints and correlations. In: Schön I, Martens K, Dijk P (eds) _Lost sex._ Springer: Netherlands, pp 447–474 Chapter Google Scholar * Korchagin VI, Badaeva TN, Tokarskaya
ON, Martirosyan IA, Darevsky IS, Ryskov AP (2007) Molecular characterization of allelic variants of (GATA)n microsatellite loci in parthenogenetic lizards _Darevskia unisexualis_
(Lacertidae). _Gene_ 392:126–133 Article CAS PubMed Google Scholar * Kupriyanova L (2009) Cytogenetic and genetic trends in the evolution of unisexual lizards. _Cytogenet Genome Res_
127:273–279 Article CAS PubMed Google Scholar * Lehmann GUC, Siozios S, Bourtzis K, Reinhold K, Lehmann AW (2011) Thelytokous parthenogenesis and the heterogeneous decay of mating
behaviours in a bushcricket (Orthopterida). _J Zool Syst Evol Res_ 49:102–109 Article Google Scholar * Lively CM (2010) A review of red queen models for the persistence of obligate sexual
reproduction. _J Hered_ 101:S13–S20 Article PubMed Google Scholar * Luijckx P, Ho EKH, Gasim M, Chen S, Stanic A, Yanchus C et al. (2017) Higher rates of sex evolve during adaptation to
more complex environments. _Proc Natl Acad Sci USA_ 114:534–539 Article CAS Google Scholar * Lutes AA, Baumann DP, Neaves WB, Baumann P (2011) Laboratory synthesis of an independently
reproducing vertebrate species. _Proc Natl Acad Sci_ _USA_ 108:9910–9915 Article CAS Google Scholar * Maynard Smith J (1978) _The evolution of sex._ Cambridge University Press: New York,
NY Google Scholar * McDonald MJ, Rice DP, Desai MM (2016) Sex speeds adaptation by altering the dynamics of molecular evolution. _Nature_ 531:233–236 Article CAS PubMed PubMed Central
Google Scholar * Meier J, Marques D, Mwaiko S, Wagner CE, Excoffier L, Seehausen O (2017) Ancient hybridization fuels rapid cichlid fish adaptive radiations. _Nat Commun._ 8:14363 * Moritz
C, Brown WM, Densmore LD, Vyas S, Donnellan S, Adams M et al. (1989) Genetic diversity and the dynamics of hybrid parthenogenesis in _Cnemidophorus_ (Teiidae) and _Heteronotia_ (Gekkonidae).
In: Dawley RM, Bogart JP (eds) _Evolution and ecology of unisexual vertebrates_. New York State Museum: Albany * Moritz C, Donnellan S, Adams M, Baverstock PR (1989) The origin and
evolution of parthenogenesis in _Heteronotia binoei_ (Gekkonidae): extensive genotypic diversity among parthenogens. _Evolution_ 43:994–1003 Article CAS PubMed Google Scholar * Moritz C,
Uzzell T, Spolsky C, Hotz H, Darevsky I, Kupriyanova L et al. (1992) The maternal ancestry and approximate age of parthenogenetic species of Caucasian rock lizards (Lacerta: Lacertidae).
_Genetica_ 87:53–62 Article CAS Google Scholar * Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA (2006) Clonal origin and evolution of a transmissible. _Cancer Cell_ 126:477–487
Article CAS PubMed PubMed Central Google Scholar * Murphy R (2000) A fine line between sex and unisexuality: the phylogenetic constraints on parthenogenesis in lacertid lizards. _Zool J
Linn Soc_ 130:527–549 Article Google Scholar * Murphy RW, Darevsky IS, MacCulloch RD, Fu J, Kupriyanova LA (1996) Evolution of the bisexual species of Caucasian rock lizards: a
phylogenetic evaluation of allozyme data. _Russ J Herpetol_ 3:18–31 * Otto SP (2009) The evolutionary enigma of sex. _Am Nat_ 174:S1–S14 Article PubMed Google Scholar * Pinho C, Sequeira
F, Godinho R, Harris DJ, Ferrand N (2004) Isolation and characterization of nine microsatellite loci in _Podarcis bocagei_ (Squamata: Lacertidae). _Mol Ecol Notes_ 4:286–288 Article CAS
Google Scholar * Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. _Genetics_ 155:945–959 * Reeder TW, Cole CJ, Dessauer HC
(2002) Phylogenetic relationships of whiptail lizards of the genus _Cnemidophorus_ (Squamata: Teiidae): a test of monophyly, reevaluation of karyotypic evolution, and review of hybrid
origins. _Am Mus Novit_ 3365:1–61 Article Google Scholar * Remón N, Vila M, Galán P, Naveira H (2008) Isolation and characterization of polymorphic microsatellite markers in _Iberolacerta
monticola_, and cross-species amplification in _Iberolacerta galani_ and _Zootoca vivipara_. _Mol Ecol Resour_ 8:1351–1353 Article PubMed CAS Google Scholar * Rice WR (1989) Analyzing
tables of statistical tests. _Evolution_ 43:223–225 Article PubMed Google Scholar * Ryskov AP, Osipov FA, Omelchenko AV, Semyenova SK, Girnyk AE, Korchagin VI et al. (2017) The origin of
multiple clones in the parthenogenetic lizard species Darevskia rostombekowi. _PLoS ONE_12:e0185161 Article PubMed PubMed Central CAS Google Scholar * Sambrook J, Russell DW (2001)
_Molecular cloning: a laboratory manual_. Cold Spring Harbor Laboratory Press * Schwander T, Crespi BJ (2009) Multiple direct transitions from sexual reproduction to apomictic
parthenogenesis in _Timema_ stick insects. _Evol Int J Org Evol_ 63:84–103 Article PubMed Google Scholar * Sinclair EA, Pramuk JB, Bezy RL, Crandall KA, Sites JW Jr (2010) DNA evidence
for nonhybrid origins of parthenogenesis in natural populations of vertebrates. _Evolution_ 64:1346–1357 * Sillero N, Argaña E, Freitas S, García-Muñoz E, Arakelian M, Corti C et al. (2018)
Short term spatial structure of a lizard (_Darevskia_ sp.) community in Armenia. _Acta Herpetol_ 13:155–163 * Shine R, Phillips B, Langkilde T, Lutterschmidt DI, Waye H, Mason RT (2004)
Mechanisms and consequences of sexual conflict in garter snakes (_Thamnophis sirtalis_, Colubridae). _Behav Ecol_ 15:654–660 Article Google Scholar * Tarkhnishvili D, Gavashelishvili A,
Avaliani A, Murtskhvaladze M, Mumladze L (2010) Unisexual rock lizard might be outcompeting its bisexual progenitors in the Caucasus. _Biol J Linn Soc_ 101:447–460 Article Google Scholar *
Tarkhnishvili D, Murtskhvaladze M, Anderson CL (2017) Coincidence of genotypes at two loci in two parthenogenetic rock lizards: how backcrosses might trigger adaptive speciation. _Biol J
Linn Soc_ 121:365–378 Article Google Scholar * Tarkhnishvili D, Murtskhvaladze M, Gavashelishvili A (2013) Speciation in Caucasian lizards: climatic dissimilarity of the habitats is more
important than isolation time. _Biol J Linn Soc_ 109:876–892 Article Google Scholar * Taylor HL, Walker JM, Cole CJ, Dessauer HC (2015) Morphological divergence and genetic variation in
the triploid parthenogenetic teiid lizard, _Aspidoscelis neotesselata_. _J Herpetol_ 49:491–501 Article Google Scholar * Trifonov VA, Paoletti A, Barucchi VC, Kalinina T, O’Brien PCM,
Ferguson-Smith MA et al. (2015) Comparative chromosome painting and NOR distribution suggest a complex hybrid origin of triploid _Lepidodactylus lugubris_ (Gekkonidae). _PLoS ONE_
10:e0132380 Article PubMed PubMed Central CAS Google Scholar * van der Kooi CJ, Schwander T (2014) On the fate of sexual traits under asexuality. _Biol Rev_ 89:805–819 Article PubMed
Google Scholar * Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) micro-checker: software for identifying and correcting genotyping errors in microsatellite data. _Mol Ecol
Notes_ 4:535–538 Article CAS Google Scholar * Vergun AA, Martirosyan IA, Semyenova SK, Omelchenko AV, Petrosyan VG, Lazebny OE et al. (2014) Clonal diversity and clone formation in the
parthenogenetic Caucasian rock lizard _Darevskia dahli_. _PLoS ONE_ 9:e91674 Article PubMed PubMed Central CAS Google Scholar * Weismann A (1889) The significance of sexual reproduction
in the theory of natural selection. In: Poulton EB, Shipley AE (eds) _Essays upon heredity and kindred biological problems_. Clarendon Press: Oxford, p 251–332. * Wetherington JD, Kotora
KE, Vrijenhoek RC (1987) A test of the spontaneous heterosis hypothesis for unisexual vertebrates. _Evolution_ 41:721–731 Article PubMed Google Scholar * Wellenreuther M, Runemark A,
Svensson EI, Hansson B (2009) Isolation and characterization of polymorphic microsatellite loci for the Skyros wall lizard _Podarcis gaigeae_ (Squamata: Lacertidae). _Mol Ecol Resour_
9:1005–1008 Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by the project “Preserving Armenian biodiversity: Joint Portuguese – Armenian
program for training in modern conservation biology” of Gulbenkian Foundation (Portugal) and the FCT grant SFRH/BD/81483/2011. The authors thank Anna Vardanyan, Claudia Corti and Elena
Argaña for field assistance, Andy Krupa, Gavin Horsburgh and Susana Lopes for their assistance with the genotyping, and Steeves Buckland and Óscar Mira for their suggestions in the methods.
We also thank Karel Janko and an anonymous reviewer for comments on previous versions of the manuscript. MA was supported by SCS MES RA - RFFR 18 RF-132 project. Capture permits were
provided by the Ministry of Nature Protection of Republic Armenia Code 5/22.1/51043 and lizard handling followed the ethical guidelines of Yerevan State University. AUTHOR INFORMATION Author
notes * Susana N. Freitas Present address: Department of Ecology and Evolution, University of Lausanne, Lausanne, Switzerland * These authors contributed equally: Roger K. Butlin, Miguel A.
Carretero AUTHORS AND AFFILIATIONS * Department of Animal and Plant Sciences, The University of Sheffield, Sheffield, S10 2TN, UK Susana N. Freitas & Roger K. Butlin * CIBIO Research
Centre in Biodiversity and Genetic Resources, InBIO, Universidade do Porto, Campus de Vairão, Rua Padre Armando Quintas, N° 7. 4485-661 Vairão, Vila do Conde, Portugal Susana N. Freitas, D.
James Harris & Miguel A. Carretero * CICGE Centro de Investigação em Ciências Geo-Espaciais, Faculdade de Ciências da Universidade do Porto, Observatório Astronómico Prof. Manuel de
Barros, Alameda do Monte da Virgem, 4430-146, Vila Nova de Gaia, Portugal Neftalí Sillero * Yerevan State University, Alek Manoogian, 1, 0025, Yerevan, Armenia Marine Arakelyan * Department
of Marine Sciences, Universitsy of Gothenburg, 405 30, Gothenburg, Sweden Roger K. Butlin Authors * Susana N. Freitas View author publications You can also search for this author inPubMed
Google Scholar * D. James Harris View author publications You can also search for this author inPubMed Google Scholar * Neftalí Sillero View author publications You can also search for this
author inPubMed Google Scholar * Marine Arakelyan View author publications You can also search for this author inPubMed Google Scholar * Roger K. Butlin View author publications You can also
search for this author inPubMed Google Scholar * Miguel A. Carretero View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence
to Susana N. Freitas. ETHICS DECLARATIONS CONFLICT OF INTEREST The authors declare that they have no conflict of interest. ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY MATERIAL FIGURE S1 FIGURE S2 FIGURE S4 FIGURE S5 FIGURE
S6 FIGURE S7 FIGURE S8 FIGURE S3 TABLE S1 TABLE S2 TABLE S3 TABLE S4 TABLE S5 TABLE S6 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Freitas, S.N.,
Harris, D.J., Sillero, N. _et al._ The role of hybridisation in the origin and evolutionary persistence of vertebrate parthenogens: a case study of _Darevskia_ lizards. _Heredity_ 123,
795–808 (2019). https://doi.org/10.1038/s41437-019-0256-5 Download citation * Received: 05 November 2018 * Revised: 12 June 2019 * Accepted: 18 June 2019 * Published: 14 August 2019 * Issue
Date: December 2019 * DOI: https://doi.org/10.1038/s41437-019-0256-5 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry,
a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative