People from ibiza: an unexpected isolate in the western mediterranean
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT In this study, we seek to understand and to correlate the genetic patterns observed in the population of the island of Ibiza in the Western Mediterranean basin with past events.
Genome-wide genotypes of 189 samples representing 13 of 17 regions in Spain have been analyzed, in addition to 105 samples from the Levant, 157 samples from North Africa, and one ancient
sample from the Phoenician Cas Molí site in Ibiza. Before the Catalans conquered the island in 1235 CE, Ibiza (_Eivissa_) had already been influenced by several cultures, starting with the
Phoenicians, then the Carthaginians, followed by the Umayyads. The impact of these various cultures on the genetic structure of the islanders is still unexplored. Our results show a clear
distinction between Ibiza and the rest of Spain. To investigate whether this was due to the Phoenician colonization or to more recent events, we compared the genomes of current Ibizans to
that of an ancient Phoenician sample from Ibiza and to both modern Levantine and North African genomes. We did not identify any trace of Phoenician ancestry in the current Ibizans.
Interestingly, the analysis of runs of homozygosity and changes in the effective population size through time support the idea that drift has shaped the genetic structure of current Ibizans.
In addition to the small carrying capacity of the island, Ibiza experienced a series of dramatic demographic changes due to several instances of famine, war, malaria and plague that could
have significantly contributed to its current genetic differentiation. You have full access to this article via your institution. Download PDF SIMILAR CONTENT BEING VIEWED BY OTHERS 9,000
YEARS OF GENETIC CONTINUITY IN SOUTHERNMOST AFRICA DEMONSTRATED AT OAKHURST ROCKSHELTER Article Open access 19 September 2024 THE GENOMIC LANDSCAPE OF MEXICAN INDIGENOUS POPULATIONS BRINGS
INSIGHTS INTO THE PEOPLING OF THE AMERICAS Article Open access 12 October 2021 PUNIC PEOPLE WERE GENETICALLY DIVERSE WITH ALMOST NO LEVANTINE ANCESTORS Article 23 April 2025 INTRODUCTION
Island populations tend to be small and isolated, and thus prone to the effects of genetic drift, particularly through bottlenecks and founder effects. The outcome is represented by reduced
genetic diversity, extended identity by descent (IBD) sharing, and a higher number of runs of homozygosity (ROHs) [1]. The loss of genetic diversity after a sudden decrease in population
size is a process that can be more easily seen in environments that have experienced geographical and/or cultural isolation. Culturally speaking, islands represent both shelters for the
conservation of traditions, and innovative places for driving processes of social change [2]. In the landscape of the Western Mediterranean, the complex history of Ibiza stands out for its
peculiarity. Ibiza (_Eivissa_ in Catalan) is one of the Balearic Islands, the closest to the continent (~90 Km away) but also the most arid. The island has an area of 577 Km [2], and, until
the 1960s, its population never exceeded 35,000 inhabitants, while currently it is ~140,000 (ref. [3]). Archeological evidence suggests that the island was sporadically visited from the late
6th millennium to the early 5th millennium BCE but was probably not permanently settled until much later. No further evidence of human presence is found until the late 3rd millennium BCE,
in the Neolithic/Calcolithic. The Bronze Age, megalithic _talaiòtic_ culture of Mallorca and Minorca is absent in Ibiza, probably due to the low carrying capacity of the island resulting in
a sparse population density. This also explains how the Phoenicians could settle in Ibiza with little apparent resistance [4]. The Phoenician _Ibossim_ was founded in 654 BCE and, given the
scarcity of pre-Phoenician sites discovered to date, it is suggested that the Phoenicians were the first permanent settlers of the island [5]. Around 550 BCE the Carthaginians, who were the
descendants of the Phoenician founders of Carthage, replaced the early Phoenicians settlers [6] and inhabited the island for the next five centuries, changing the name of the island to
_Ebusus_. In 123 BCE, the island was annexed to the Roman Republic, but maintained its cultural and political independence [5]; it was after the foundation of the Roman Empire in 27 BCE that
the island lost its independence to become a federate city [6]. Later, around 902 CE the Umayyads of Cordoba occupied the island for about four centuries and Ibiza became _Yebisah_ [6],
going through a process of islamization, as suggested by archaeological records [7]. In 1235, the invasion of the Catalans represented the last significant event in the colonization history
of the island before it became the privileged destination of extensive immigration from the _hippy_ community in the second half of the 20th century, paving the way for Ibiza to become the
cosmopolitan island it is today. All of these events must have contributed in shaping the genetic background of Ibizans, but their impact on the modern islanders’ genome remains unknown. A
previous study based on classical genetic markers proposed a genetic continuity of the islanders with North African and East Mediterranean populations [5]. A Carthaginian maternal founder
effect was also hypothesized based on analyses of the mitochondrial hypervariable region I (HVRI) [8]. However, no reduction of diversity was detected for Y-chromosome STRs, as compared with
other Mediterranean groups [9]. In this study we aim to explore genomic diversity by conducting the first genome-wide analysis of the modern Ibizan population. We also aim to investigate
whether Phoenician ancestry can be identified in modern Ibizans or whether this ancestry was lost due to demographic replacement and subsequent isolation. MATERIALS AND METHODS DATASET AND
GENOTYPES Informed consent was given by 163 volunteers from three regions in Spain (Catalonia, Valencia, and the Balearic Islands; as a shorthand, we will refer to these as the _core
regions_ of our study); Internal Review Board approval for this work was granted by CEIC-PSMAR ref. 2016/6723/I. This research was conducted under the principles of the Helsinki declaration.
The geographical origin of each subject was characterized and recorded as the average coordinates of the birthplaces of their four grandparents, which were distant at most 50 Km. For some
analyses, a finer-grained geographical scale was used (by island in the Balearic Islands, by province in Valencia, and _regió_, namely the first-level administrative division used by the
Catalan government, plus the city of Barcelona in Catalonia). DNA samples were collected from saliva and extracted as described in Solé-Morata et al. [10]. Overall, four Axiom®Genome-Wide
Human Origins Arrays (~629 K SNPs) [11] were genotyped at the Centro Nacional de Genotipado - Universidade de Santiago de Compostela facility. Genotype calling was performed applying the
Affymetrix Best Practices Workflow implemented in the software AxiomTM Analysis Suite 2.0. Since the samples were distributed in different plates, two separate batches were run to avoid any
batch effect during the genotype calling. A total of 41 samples failed the genotyping process, leading to 122 remaining individuals (Supplementary Table S1). An Identity-by-descent (IBD)
analysis was performed in order to unveil close relatedness between the donors. Two pairs of samples, both from the island of Ibiza, did not pass the test. Eventually, a total of 120 samples
were retained (data available at https://figshare.com/articles/People_from_Ibiza_dataset/7162025). Out of the 633,995 SNPs interrogated by the array, a different number of variants per
batch passed the genotyping process (Supplementary Figure S1). After the genotype calling, and before the quality control filtering, the initial overlap between the four plates included
569,999 SNPs. The latter decreased to 519,297 variants when a further 69 Spanish samples from freely available data [12] were added. Thus, the final dataset increased to 189 samples,
covering 13 out of the 17 regions in Spain (Supplementary Figure S2). An ancient sample (MS10614) from the Phoenician necropolis of Cas Molí, on Ibiza, was included in the work. Genome-wide
data for this sample were produced as described in Zalloua et al. [13]. Using 105 modern samples [12] from Lebanon, Syria, Israel, and Jordan, we reconstructed the ancient region of Canaan
that we then merged together with our subjects from the core regions. Further 157 subjects [14] from Morocco, Algeria, and Tunisia were used for testing North African ancestry. DATA QUALITY
CONTROL WITH PLINK X-chromosome and uniparental markers were excluded using the flag --not-chr implemented in PLINK [15] 1.90. Samples showing more than 10% of SNP genotypes missing were
removed. We also rejected SNPs missing in more than 95% of the samples, and SNPs with extreme deviation from the Hardy-Weinberg equilibrium (_p_ < 10−5), which left 519,249 SNPs; removal
of SNPs displaying little variation in the sample set (MAF < 0.05) resulted in 348,443 remaining SNPs. A set of independent SNPs was generated by pruning for linkage disequilibrium using
the --indep-pairwise flag (the window size was set to 200 SNPs with a shifting of 25 SNPs, and _r_2 set to 0.5). We then performed an Identity-by-descent (IBD) analysis using the flag
--genome in order to detect those samples with cryptic relatedness. A cutoff was applied up to the third degree of relationship (PI_HAT > 0.125), and for each related couple of samples
the one showing the lowest SNP call rate was removed. Only the common SNPs to all the merged datasets were retained. After the quality control, polymorphism and LD filters in the global
dataset of 189 samples, 142,520 common SNPs passed and were used in further analyses. A different number of variants were achieved when considering the dataset with 120 samples from the
three core regions (151,286 SNPs remaining after QC). In the analysis with the North African subjects, 55,969 final variants were used, while 47,057 SNPs were retained in the analysis that
included the Levantine regions and the ancient sample. PRINCIPAL COMPONENT ANALYSIS (PCA) The SmartPCA program (v. 13050) in EIGENSOFT [16] (v. 6.0.1) was used for computing the
eigenvectors, and the results were plotted in R (v. 3.0.1). FIXATION INDEX (_F_ _ST_) The pairwise differentiation was computed between the different groups relying on the Weir and
Cockerham’s _F__ST_ measure [17] implemented in the R package StAMPP [18]. Using the function stamppFst, we bootstrapped across loci (1000 repetitions) in order to estimate the P-values
associated to each _F__ST_ measure. Also, a percentile method was implemented in order to compute the confidence intervals; a standard value of 95% was set to define the upper and the lower
confidence intervals. Heatmaps were produced in RStudio [19] using R version 3.4.4 [20]. The Canary Islands sample was not included in this and all subsequent analyses given its extremely
low sample size (_N_ = 2). MANTEL TEST A geographic distance matrix was produced using The Geographic Distance Matrix Generator (v. 1.2.3, available from
http://biodiversityinformatics.amnh.org/open_source/gdmg) and, together with the _F__ST_ matrix, was used as input for the Mantel test calculation using the ade4 (ref. [21]) library in R.
The plot was also generated with R using both reshape [22] and ggplot2 (ref. [23]) libraries. ADMIXTURE Individual ancestries were computed with ADMIXTURE (v. 1.3.0) [24] and tested from _K_
= 2 up to _K_ = 8. A consensus for every _K_ was calculated out of 10 independent runs using CLUMPP [25] (v. 1.1.2), while Distruct [26] (v. 1.1) was used for plotting the results for each
one of the Ks. Furthermore, for _K_ = 2 up to _K_ = 5, each component was individually displayed along the geographic coordinates of the samples, resulting in the geographic distribution of
the admixture percentages. These latter plots were obtained using the R function _tessplot_ implemented in the plot.admixture.r script (freely available at
http://membres-timc.imag.fr/Olivier.Francois/plot.admixture.r). Admixture f3 statistics were computed using the threepop function implemented in TreeMix [27]. RUNS OF HOMOZYGOSITY (ROH)
Pruned data underwent ROH analysis performed in PLINK. We scanned the genome using sliding windows containing at least 50 SNPs and a minimum length of 500 kb. At most, 1 heterozygous call
and 5 missing calls per window were allowed. A threshold of 0.05 was set to define the proportion of homozygous windows overlapping the position of each SNP; at least one SNP per 50 kb was
chosen as required minimum density value, and two consecutive SNPs were included in the same ROH when a maximum gap of 100 kb split them up. For each group, the average ROH number was
calculated dividing the total number of runs (NROH) by the number of samples. The average total ROH length (namely, the average SROH per sample in each group) was calculated as the total ROH
length (SROH) divided by the total number of individuals. The FROH (amount of autosomal genome covered by ROHs) for runs greater than 1 Mb was calculated according to the formula presented
in McQuillan et al. [28]: $${\rm{FROH}}_{ > 1{\rm{Mb}}} = \frac{{{\sum } {\mathrm{L}}_{{\mathrm{ROH}}}}}{{{{\rm{L}}}_{{\mathrm{auto}}}}}$$ (1) In this formula, \(\mathop {\sum
}\nolimits^{} {\mathrm{L}}_{{\mathrm{ROH}}}\) is the SROH>1Mb calculated for each individual, and \({{L}}_{{\mathrm{auto}}}\) is a factor representing the length of the autosomal genome
covered by SNPs on the array data (2,782.7 Mb). IBDSEQ/IBDNE In order to detect pairwise IBD blocks, we ran the program IBDseq [29] with default settings for the global dataset of 189
samples. We then extracted from the output the results for each group and used those ones as input for IBDNe [30] program. The latter allowed us to estimate the variation of _N_e through the
past few generations since we defined longer IBD segments by setting a cutoff at 7 cM (minibd=7). Apart from the minibd option that we customly set, we ran IBDNe with default settings,
filtering the IBD segments as suggested by Browning & Browning [30]. Also, according to the software settings, 50 generations is the most reliable range we can take into account when
working with SNP array data. We set a threshold of 100 IBD segments as the minimum sample size for this analysis, which was met only by the Ibizan and Catalan samples (440 and 223,
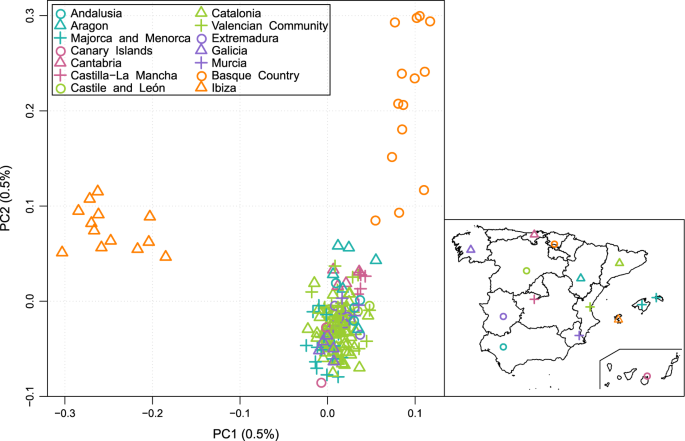
respectively). RESULTS GENETIC DIFFERENTIATION IN A SPANISH CONTEXT A first Principal Component Analysis (PCA) was performed considering the entire dataset of 189 Spanish samples (120 new
plus 69 previously published). In the plot (Fig. 1), the first PC clearly separates all of the Ibizan samples from the rest of the Spanish subjects, while the second PC separates Basque
samples from the rest, within which no structure can be discerned. Because PC1 and PC2 explain approximately the same proportion of variance [31], these results imply that the genetic
differentiation of Ibiza from the rest of Spain is at least of the same order of magnitude as that of the Basque Country. When we narrowed the analysis down to 120 samples in the three core
regions (Catalonia, Valencia, and the Balearic Islands) we still detected two separate clusters defined along the first PC (Supplementary Figure S3). Next, we quantified the genetic
variation between groups through _F__ST_. Apart from Ibiza and the Basque Country, _F_ST ranged from 4 × 10−5 to 0.002, while population pairs containing Ibiza had _F__ST_ values ranging
from 0.006 to 0.01 (Supplementary Figure S4a and Table S2). Within the core regions, _F__ST_ was even more homogeneous, ranging from 3 × 10−6 to 0.002, while Ibiza showed higher values of
differentiation (0.006–0.007, Supplementary Figure S4b). A Mantel test between the _F_ST values and the geographic distances showed a positive but not significant correlation for both the
global data set (_R_2 = 0.01, _P_ = 0.4), and within the core regions (_R_2 = 0.164, _P_ = 0.2). Interestingly, when Ibiza was excluded from the test, the positive correlation became
significant in the core regions, with _R_2 = 0.353 and _P_ = 0.009 (Supplementary Figure S5a). This is likely a reflection of the genetic outlier position of Ibiza, since its differentiation
is larger than expected based on its geographical position, to the point that its presence in the analysis dramatically decreases the correlation that exists between genetic and geographic
distances when Ibiza is excluded. On the other hand, the global dataset maintained a _p_ value > 0.05 (Supplementary Figure S5b), which can be attributed to the presence of another
genetic outlier, namely the Basques. In order to determine the global ancestry components of our samples, an ADMIXTURE analysis was performed, in which we found that _K_ = 2 presented the
lowest cross-validation error. The two major components detected (Fig. 2) mirror the PCA results: one component prevails in Ibiza, the other in the Basque Country, and all other individuals
contain on average 31.78% of the first vs. 68.22% of the second. Notably, moving through the different numbers of theoretical ancestral populations tested, we see a consistent pattern in the
fraction of the component coming from Ibiza (with an average of 87.45% along the eight Ks tested), while the other source starts to break up at _K_ = 3, as it is shown in the Admixture-map
plots (_K_ = 2 up to _K_ = 5 shown in Supplementary Figure S6). COMPARISON WITH AN ANCIENT PHOENICIAN SAMPLE A sample (MS10614) from the Phoenician Cas Molí site in Ibiza [13] was sequenced
at a 0.47X coverage, and was projected into a PCA space with two modern groups of samples, from Iberia (including contemporary Ibiza) and the Levant (Supplementary Figure S7a). The ancient
Phoenician sample clusters with the modern Levantines, rather than with the cluster represented by the core regions. Furthermore, the admixture proportions seem to be consistent with this
result (Supplementary Figure S7b). In the figure, the average percentage for each component is displayed for each group; it is clear that the ancient sample is closely related to the groups
from Levant. And, conversely, these results seem to be incompatible with direct continuity between the current population of Ibiza and the ancient Phoenicians who settled the island.
ADMIXTURE INTO IBIZA When we tested Ibiza for both East Mediterranean and North African ancestries, we did not find any major signal of differentiation from the rest of the Spanish groups.
In the admixture analysis with the 105 Levantine subjects, _K_ = 2 turned out to be the most suitable model according to the cross-validation error analysis; the two components we detected
can be clearly attributed to the Iberian Peninsula and the Levant, respectively (Supplementary Figure S7b). Concerning the North African influence, a principal component analysis clearly
shows a separation of the three core regions from the North African samples (Supplementary Figure S8a). This split is also corroborated by the admixture results (Supplementary Figure S8b);
even in this case, _K_ = 2 describes the structure of the groups highlighting the presence of two distinct components. As in the PCA, Ibiza conforms to the rest of the Spanish groups. The
absence of any signal of admixture in the Ibizan population was also supported by f3 statistics; in Supplementary Figure S9 we show the results for all the tested trios, including Northern
Africans, Middle Easterners, and also the ancient sample from Ibiza. ENDOGAMY AND ISOLATION IN IBIZA Endogamy in small population isolates and inbreeding leave their mark in the genome as
long runs of homozygosity (ROHs). When we separated the ROHs according to different length categories as in Kirin et al. [32], it was clear that Ibiza stood out for both average ROH number,
and average total ROH length for the interval 1–2 Mb (Figs. 3a and 3b), and was second only to Cantabria (with a huge dispersion) in the interval 2–4 Mb. Possibly, the scarce number of
samples for some groups in this category of ROH can explain this contingency; indeed, the proportion of individuals in Ibiza carrying ROHs in this size range is 76%, while it is at most 50%
in any other population (Fig. 3c). The observed ROH configuration in Ibiza seems to fit the very recent history of modern islanders; short ROHs are common in populations that have
experienced a bottleneck, reflecting also a more recent parental relatedness [33]. In our analysis, Ibiza shows the highest levels of the total length of ROHs (SROH, Supplementary Figure
S10a) with the subjects showing a homogeneous distribution through the different cumulative lengths, while different groups show outliers with values well further away from the mean. As for
the total number of ROHs (NROH), Ibiza is second only to the Basque Country (Supplementary Figure S10b), as we can also appreciate in Figure S11a in which the Ibizan samples are more widely
spread along the two axes than any other group. On the other hand, the majority of the other groups mass in an area of the plot represented by fewer and shorter ROHs. Interestingly, samples
from the Basque Country show an intermediate distribution along the axes, with some samples from the Catalan group spread through higher values, mirroring the same pattern observed for the
outliers seen in the whisker plots (for a clearer visualization, refer to Supplementary Figure S11b)). Furthermore, in Supplementary Figure S10c it is possible to appreciate how the subjects
from Ibiza stand out from the others for the proportion of the autosomal genome covered by runs of homozygosity (FROH). On average, each subject from Ibiza displays 0.81% of the genome as
autozygous, the highest value retrieved among all the groups (Supplementary Table S3). DEMOGRAPHIC HISTORY By using IBDseq and IBDNe, we calculated a series of effective population size
(_N_e) values through the last 50 generations from the present. Out of all the pairwise IBD segments retrieved with IBDseq, only a limited number passed the filter for minimum allowed length
set in IBDNe (minibd = 7 cM), with Ibiza showing the highest fraction of passing IBD segments (20.27%). _N_e through time is plotted in Fig. 4; for Ibiza, the general trend in the last 1500
years is clearly a reduction of the effective population size, up to a minimum Ne found 10-15 generations in the past, followed by an exponential growth in more recent times. When comparing
this pattern with that found in the mainland Catalan population, we detected a general similar trend but higher _N_e values, and a faster increase of the slope after the observed minimum.
We used the IBDseq output in order to quantify the amount of IBD chunks that each sample from Ibiza receives on average from the other groups (Supplementary Figure S12a). In this analysis we
subsampled both Catalan and Valencian groups in order to avoid any sample size bias. We found that each subject from Ibiza receives the highest amount of IBD chunks from its own group.
Furthermore, we calculated the total length (bp) of shared IBD chunks, and represented how much on average, each Ibizan sample receives from every other population (Supplementary Figure
S12b); the results highlight that Ibizans receive on average the longest IBD chunks from the Ibizan group itself. DISCUSSION Our study suggests that the population of Ibiza is genetically
distinct from the rest of Spain, a similar phenomenon to what has been observed with the Basques who have been previously described as a genetic outlier [34, 35, 36]. The different
approaches we applied in this study support the fact that the island is a genetic isolate. The results we achieved actually reflect the same pattern observed for other known isolated
populations. For example, the principal component analysis clearly points out that Ibiza forms its own cluster, recalling the way Sardinia splits from the rest of the Italian Peninsula [37].
This separation is also supported by the genetic variation analysis between groups. Indeed, even if a general homogeneity is detected, Ibiza shows higher values of differentiation.
Following the parallelism with Sardinia, the Italian island diverges from other European groups for _F_ST values [34] that, interestingly, mirror the ones that we observed when comparing
Ibiza to the different regions of Spain. Furthermore, it is of particular interest to notice that the value detected for the genetic distance of Ibiza from the Basque Country recalls the one
that Sardinia shares with that particular community (_F__ST_ = 0.011) [34]. The latter observation was also supported by a further _F__ST_ analysis (data not shown) including Sardinian
samples genotyped with the Human Origins Array from Lazaridis et al. [12]; the results were similar to those observed in previous studies [34]. Even when we look at the admixture results,
the pattern displayed at _K_ = 2 underscores the presence of a strong signature component that Ibiza preserves through all the K values we tested, without showing any significant break. This
is different from the behavior displayed by any of the other groups. The nature of this signature component as a signal of isolation seems to be a very common pattern observed in other
genetic isolates, such as the Kalash [38], and the Sardinians [39]. Furthermore, the configuration observed for both the Ibizan and the Basque components from _K_ = 3 on, could be the result
of a genetic drift-driven redistribution of the allele frequencies, which seems to be common in populations that experienced a rapid size reduction [40]. Since genetic differentiation can
be the consequence of very different events, we tested various hypotheses for the reasons behind the differentiation of modern Ibizans. GEOGRAPHICAL ISOLATION The hypothesis that modern
Ibizans differ from continental Spanish populations just because they live far from them has been taken into account. A Mantel test highlighted a significant correlation between the
geographical distances and the genetic ones, only when Ibiza was removed from the analysis (_R_2 = 0.353, _P_ = 0.009). This can be taken as an indication of the genetic outlier position of
Ibiza given that its presence breaks the correlation between genetic and geographic distances; note that this analysis also contains the other Balearic Islands (Mallorca and Minorca), the
latter of which is similar in area and population size to Ibiza, and is further away from the mainland. Thus, the insularity and geographical position of Ibiza do not seem to be major
contributors to its genetic distinctiveness. ARE IBIZANS DESCENDED FROM THE PHOENICIAN SETTLERS IN THE ISLAND? We tested whether Phoenicians, represented by an ancient DNA, low-coverage
genome, had contributed to the genetic make-up of present-day islanders, and tested signals of admixture with modern Eastern Mediterranean and North African populations. All the
archeological evidence to date suggest that it was the Phoenicians who first settled Ibiza, giving it a central role in the Western Mediterranean trade network, and making it “one of the
true cities of the Western European antiquity” [6]. With five centuries of colonization, it has long been suggested that the legacy of the ancient Phoenicians might be contained in the
genomes of current Ibizans [5, 8, 9, 41]. We have had the opportunity to properly explore this possibility because of the availability of an ancient Phoenician genome from Ibiza. As pointed
out by the results, it is evident that there is no genetic continuity between the modern inhabitants of the island and this ancient subject. Furthermore, our results exclude any major Middle
Eastern specific component in modern Ibizans, or any other Spanish population. Nevertheless, it is surprising how the modern Levantines still share a genetic continuity with the ancient
Ibizan (MS10614), pointing to the possibility that modern samples from those regions might be considered as proxies of the founding civilization. Also, a greater similarity to the rest of
the Iberians rather than to Levantines in this analysis suggests that the modern Ibizan differentiation is not caused by any other Ibizan-specific external admixture contribution. In support
of this observation, our results also exclude any major North African component in modern Ibizans, as well in the rest of the Spanish subjects. Also, testing more specifically for
admixture, the f3 statistics gave no signal of admixture when we tested the Ibizan group against Northern Africans, Middle Easterners, and the ancient sample. According to historical and
archaeological records, apart from the European ones, Ibiza owes its ancestry to East Mediterranean and North African populations [5]; our results support the idea that, even if any external
influence occurred, it has not persisted to the current Ibizan population. IBIZA: A DEMOGRAPHIC HISTORY The exclusion of any major genetic Phoenician influence in the modern Ibizan genome
led us to explore the demographic history of the islanders considering the effects of consanguinity, given that it has long been a common practice on the island (marriages between relatives
up to the 4th degree were very frequent, and there are records of unions between second-degree cousins dated back to the late 1960s [42]). This part involved the analysis of ROHs; runs of
uninterrupted stretches of the genome, inherited identically from both parents, which can be used to unveil both recent and ancient relatedness between subjects. Normally, longer ROHs are
associated with recent inbreeding because of the lack of recombination in the short term, while shorter ROHs are symptomatic of a much older story [32]. The outcome of our analyses
highlights the presence of more, and longer ROHs in Ibiza than in any other group, a demographic scenario which is similar to the one that was previously associated to inbred populations
[28], or in general to small groups, in which the number and the length of ROHs are usually greater than those observed in larger populations [33]. Moreover, the high number of short ROHs
some of the samples show is usually symptomatic of a population bottleneck event [33]. However, the scenario depicted for the majority of the other Spanish groups is typical of large,
non-isolated populations, characterized by few and shorter ROHs [28]. Furthermore, the samples from Catalonia, Valencia, and the remaining Balearic Islands (Majorca and Menorca) present some
individuals with larger values for both NROH and SROH. These outliers, with atypically long haplotypes, may be caused by a low recombination rate in some genomic regions that has led to the
appearance of misleadingly long ROHs in outbred populations [32, 43, 44]. However, while the possibility that the haplotypes carried by these outliers could have arisen from recent parental
relatedness cannot be totally excluded, the fact that the general trend for these groups is the one usually observed in non-isolated populations seems to be the most reasonable explanation.
The Basque Country samples represent a different situation, with a combination of both shorter and longer ROHs that suggests a more complex history. On average, the subjects from Ibiza show
the highest number of ROHs for the length category 1–2 Mb, and probably for the 2–4 Mb as well (if we consider that other populations have a rather low sample size of individuals carrying
ROHs in this range). Typically, ROHs with lengths 1–2 Mb fall in a category associated to a background of parental relatedness due to limited population size [45]. Even though consanguineous
unions are not frequently observed in European populations [32, 46], in the Mediterranean area the highest percentage is recorded in Spain (3.5%) [46]. Furthermore, ROHs with lengths 2–4 Mb
have been linked to ancient parental relatedness resulting from genetic drift and frequent consanguineous practices [47]. One of the consequences of inbreeding is the inheritance of alleles
that are identical by descent, resulting in the increase of the subjects’ overall autozygosity; the higher this value is, the greater the chance that a population has experienced some sort
of inbreeding event. Our analysis on the degree of individual autozygosity (FROH) clearly pointed out that Ibizans possess the highest autosomal coverage in ROHs. Overall, our results are
consistent with the history of consanguineous unions on the island. Current Ibizans are descendants of the Catalan conquerors of the 13th century, after which the island became a
socio-geographic isolate, promoting, especially in rural areas, the spread of the relatively stable and impervious practice of marriages between relatives [48]. One of the consequences of
this habit is undoubtedly the reduction of genetic diversity and the consequent increased isonymy(decrease in the recorded variability of surname combinations [42]). Following the ROH
results, the analysis of IBD segments detected a recent reduction in the population size due to a population bottleneck. The subjects from Ibiza have shown the highest number of IBD
segments, with significant statistical support in terms of LOD scores retrieved with IBDseq. Through IBDNe, we observed how the effective population size of the island changed in the last 50
generations, with a significant collapse within the last 10-15 generations. Even if IBDNe for small datasets is prone to loss of precision in the most recent generations [30], historical
records seems to fit well with the pattern observed; the population decline we detected may correspond to a period from the second half of the 16th century to the beginning of the 18th
century; during this period, the island suffered from famine after the Franco-Ottoman attack in 1536, and was harshly impacted by the global bubonic plague epidemic in 1652 (ref. [49]). Our
results suggest a recovery in the population size beginning around the early 18th century, which is consistent with the historical record when Ibiza swore allegiance to the crown of Philip V
of Spain in 1715. Compared to Ibiza, Catalonia shows an even more rapid increase in population size, after a similar period of reduction; this too is consistent with the historical records,
since in 1651, in addition to being under siege by the Castilian troops in the Reapers’ War, a third of the population of Barcelona was devastated by the bubonic plague [49]. Moreover, in
Supplementary Figure S12a we highlight how on average each subject from Ibiza receives more IBD chunks from itself and then from the other groups, while the longest IBD chunks clearly comes
from the Ibizan group (Supplementary Figure S12b). These results support the increased genetic drift in Ibizans and their genetic isolation from other Spanish groups. However, detecting any
founder effect from the Catalan source through the analysis of the IBD sharing seems to be unlikely at a such small-scale. This is possibly due to the highly drifted Ibizan sample, and the
high homogeneity among the mainland populations. In conclusion, our study suggests that the elevated genetic differentiation Ibizans have from other Spanish groups does not result from
genetic continuity with Phoenician founders; also, the presence of more and longer ROH segments supports the hypothesis of increased parental relatedness due to the well recorded practice of
consanguineous unions on the island, while the IBD chunks distribution supports the genetic drift and also suggests that a population collapse, 10-15 generations ago, affected the island,
likely corresponding to the bubonic plague occurred in 1652. REFERENCES * Somers M, Olde Loohuis LM, Aukes MF, et al. A genetic population isolate in the Netherlands showing extensive
haplotype sharing and long regions of homozygosity. Genes. 2017;8:1–14. Article Google Scholar * Braudel F. _Une Leçon d’histoire_. 1985. Paris (France): Flammarion. * Institut
d’Estadística de les Illes Balears (IBESTAT). IBESTAT (2017). Cifras de población. https://ibestat.caib.es/ibestat/estadistiques/poblacio/padro/2acef6cf-175a-4826-b71e-8302b13c1262. * Ribas
BC Eivissa-Prehistòria. Enciclopèdia d’Eivissa i Formentera. 2006. http://www.eeif.es/veus/Eivissa-historia-prehistoria/. * Picornell A, Ana M, Castro JA, Ramon MM, Arya R, Crawfor MH.
Genetic variation in the population of ibiza (spain): genetic structure, geography, and language. Hum Biol. 1996;68:899–913. * Armstrong S. The White Island. London (UK): Transworld books.
2004. * Gurrea Barricarte R, Martín Parrilla Á. Eivissa història època califal. Enciclopèdia d’Eivissa i Formentera. 2015. http://www.eeif.es/veus/Eivissa-historia-epoca-andalusina/. *
Picornell A, Gómez-Barbeito L, Tomàs C, Castro JA, Ramon MM. Mitochondrial DNA HVRI variation in Balearic populations. Am J Phys Anthropol. 2005;128:119–30. Article CAS Google Scholar *
Tomàs C, Jiménez G, Picornell A, Castro JA, Ramon MM. Differential maternal and paternal contributions to the genetic pool of Ibiza Island, Balearic Archipelago. Am J Phys Anthropol.
2006;129:268–78. Article Google Scholar * Solé-Morata N, Bertranpetit J, Comas D, Calafell F. Y-chromosome diversity in Catalan suRNAme samples: Insights into suRNAme origin and frequency.
Eur J Hum Genet. 2015;23:1549–57. Article Google Scholar * Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y. Ancient admixture in human history. Genetics. 2012;192:1065–93.
Article Google Scholar * Lazaridis I, Patterson N, Mittnik A, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–13. Article
CAS Google Scholar * Zalloua P, Collins CJ, Gosling A, et al. Ancient DNA of Phoenician remains indicates discontinuity in the settlement history of Ibiza. Sci Rep. 2018;8:17567. Article
Google Scholar * Arauna LR, Mendoza-Revilla J, Mas-Sandoval A, et al. Recent historical migrations have shaped the gene pool of Arabs and Berbers in North Africa. Mol Biol Evol.
2017;34:318–29. CAS PubMed Google Scholar * Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet.
2007;81:559–75. Article CAS Google Scholar * Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:2074–93. Article CAS Google Scholar * Weir BS,
Clark Cockerham C. Estimating F-statistics for the analysis of population structure. Evol Vol38. 1984;38:1358–70. CAS Google Scholar * Pembleton LW, Cogan NOI, Forster JW. StAMPP: an R
package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol Ecol Resour. 2013;13:946–52. Article CAS Google Scholar * RStudio Team. RStudio
Team. RStudio: integrated development for R. 2015. http://www.rstudio.com/. * R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical
Computing. 2018. * Dray S, Dufour A-B. The ade4 Package: Implementing the duality diagram for ecologists. J Stat Softw. 2007;22. https://doi.org/10.18637/jss.v022.i04. * Wickham H. Reshaping
data with the /pkgreshape package. J Stat Softw. 2007;21:1–20. Article Google Scholar * Wickham H. ggplot2: elegant graphics for data analysis. Media. 2009;35:211. Google Scholar *
Alexander DH, Novembre J. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. Article CAS Google Scholar * Jakobsson M, Rosenberg NA. CLUMPP: a
cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–6. Article CAS Google Scholar
* Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–8. Article Google Scholar * Pickrell JK, Pritchard JK. Inference of
population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8. https://doi.org/10.1371/journal.pgen.1002967. Article CAS Google Scholar * McQuillan R,
Leutenegger AL, Abdel-Rahman R, et al. Runs of homozygosity in european populations. Am J Hum Genet. 2008;83:359–72. Article CAS Google Scholar * Browning BL, Browning SR. Detecting
identity by descent and estimating genotype error rates in sequence data. Am J Hum Genet. 2013;93:840–51. Article CAS Google Scholar * Browning SR, Browning BL. Accurate non-parametric
estimation of recent effective population size from segments of identity by descent. Am J Hum Genet. 2015;97:404–18. Article CAS Google Scholar * McVean G. A genealogical interpretation
of principal components analysis. PLoS Genet. 2009;5. https://doi.org/10.1371/journal.pgen.1000686. Article Google Scholar * Kirin M, McQuillan R, Franklin CS, Campbell H, McKeigue PM,
Wilson JF. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE. 2010;5:e13996. Article Google Scholar * Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF.
Runs of homozygosity: Windows into population history and trait architecture. Nat Rev Genet. 2018;19:220–34. Article CAS Google Scholar * Rodríguez-Ezpeleta N, Álvarez-Busto J, Imaz L,
et al. High-density SNP genotyping detects homogeneity of Spanish and French Basques, and confirms their genomic distinctiveness from other European populations. Hum Genet. 2010;128:113–7.
Article Google Scholar * Comas D, Mateu E, Calafell F, et al. HLA class I and class II DNA typing and the origin of Basques. Tissue Antigens. 1998;51:30–40. Article CAS Google Scholar *
Calafell F, Bertranpetit J. Principal component analysis of gene frequencies and the origin of Basques. Am J Phys Anthr. 1994;93:201–15. Article CAS Google Scholar * Fiorito G, Di
Gaetano C, Guarrera S, et al. The Italian genome reflects the history of Europe and the Mediterranean basin. Eur J Hum Genet. 2016;24:1056–62. Article CAS Google Scholar * Ayub Q,
Mezzavilla M, Pagani L, et al. The kalash genetic isolate: Ancient divergence, drift, and selection. Am J Hum Genet. 2015;96:775–83. Article CAS Google Scholar * Di Gaetano C, Fiorito G,
Ortu MF, et al. Sardinians genetic background explained by runs of homozygosity and genomic regions under positive selection. PLoS One. 2014;9:1–8. Google Scholar * Listman JB, Malison RT,
Sughondhabirom A, et al. Demographic changes and marker properties affect detection of human population differentiation. BMC Genet. 2007;8:21. Article Google Scholar * Adams SM, Bosch E,
Balaresque PL, et al. The genetic legacy of religious diversity and intolerance: paternal lineages of christians, jews, and muslims in the iberian peninsula. Am J Hum Genet. 2008;83:725–36.
Article CAS Google Scholar * Claudio Alarco von Perfall. Sobre los matrimonios consanguíneos en Ibiza. Eivissa, num8. 1976;16:328–31. Google Scholar * Gibson J, Morton NE, Collins A.
Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15:789–95. Article CAS Google Scholar * Christofidou P, Nelson CP, Nikpay M, et al. Runs of homozygosity:
association with coronary artery disease and gene expression in monocytes and macrophages. Am J Hum Genet. 2015;97:228–37. Article CAS Google Scholar * Pemberton TJ, Absher D, Feldman
MW, Myers RM, Rosenberg NA, Li JZ. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. 2012;91:275–92. Article CAS Google Scholar * Ceballos FC. La genética
de los matrimonios consanguíneos. Dendra Médica Rev Humanid. 2011;10:160–76. Google Scholar * Jalkh N, Sahbatou M, Chouery E, Megarbane A, Leutenegger AL, Serre JL. Genome-wide inbreeding
estimation within Lebanese communities using SNP arrays. Eur J Hum Genet. 2015;23:1364–9. Article CAS Google Scholar * Rozenberg D. Endogàmia i problemes d’identificació a Eivissa:
l’exemple de Sant Miquel. Eivissa, num. 1981;12:515–8. Google Scholar * Kohn GC. Encyclopedia of plague and pestilence: from ancient times to the present. New York (USA): Checkmark Books.
1995. Download references ACKNOWLEDGEMENTS We thank Maria Ferrer (IBE, CSIC-UPF) for her invaluable help in finding the Ibiza sample donors, and Inés Quintela (CEGEN - USC) for her
assistance in genotyping the samples. Of course, this work would not have been possible without the kind collaboration of all the sample donors. Funding was provided by the Agencia Estatal
de Investigación and Fondo Europeo de Desarollo Regional (FEDER) (grant CGL2016-75389-P), Agència de Gestió d’Ajuts Universitaris i de la Recerca (Generalitat de Catalunya) grant 2014 SGR
866, and “Unidad de Excelencia María de Maeztu”, funded by the MINECO (ref: MDM-2014-0370). SAB was supported by the Agencia Estatal de Investigación FPI grant BES-2014-069224. AUTHOR
INFORMATION AUTHORS AND AFFILIATIONS * Departament de Ciències Experimentals i de la Salut, Institute of Evolutionary Biology (CSIC-UPF), Universitat Pompeu Fabra, Barcelona, Spain Simone
Andrea Biagini, Neus Solé-Morata, David Comas & Francesc Calafell * Department of Anatomy, University of Otago, Dunedin, New Zealand Elizabeth Matisoo-Smith * School of Medicine, The
Lebanese American University, Chouran, Beirut, Lebanon Pierre Zalloua Authors * Simone Andrea Biagini View author publications You can also search for this author inPubMed Google Scholar *
Neus Solé-Morata View author publications You can also search for this author inPubMed Google Scholar * Elizabeth Matisoo-Smith View author publications You can also search for this author
inPubMed Google Scholar * Pierre Zalloua View author publications You can also search for this author inPubMed Google Scholar * David Comas View author publications You can also search for
this author inPubMed Google Scholar * Francesc Calafell View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Francesc
Calafell. ETHICS DECLARATIONS CONFLICT OF INTEREST The authors declare that they have no conflict of interest. ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer Nature remains neutral with
regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY MATERIAL RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS
ARTICLE CITE THIS ARTICLE Biagini, S.A., Solé-Morata, N., Matisoo-Smith, E. _et al._ People from Ibiza: an unexpected isolate in the Western Mediterranean. _Eur J Hum Genet_ 27, 941–951
(2019). https://doi.org/10.1038/s41431-019-0361-1 Download citation * Received: 10 August 2018 * Revised: 15 January 2019 * Accepted: 24 January 2019 * Published: 14 February 2019 * Issue
Date: June 2019 * DOI: https://doi.org/10.1038/s41431-019-0361-1 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative