Linc00426 accelerates lung adenocarcinoma progression by regulating mir-455-5p as a molecular sponge
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Increasing lines of evidence indicate the role of long non-coding RNAs (LncRNAs) in gene regulation and tumor development. Hence, it is important to elucidate the mechanisms of
LncRNAs underlying the proliferation, metastasis, and invasion of lung adenocarcinoma (LUAD). We employed microarrays to screen LncRNAs in LUAD tissues with and without lymph node metastasis
and revealed their effects on LUAD. Among them, Linc00426 was selected for further exploration in its expression, the biological significance, and the underlying molecular mechanisms.
Linc00426 exhibits ectopic expression in LUAD tissues and cells. The ectopic expression has been clinically linked to tumor size, lymphatic metastasis, and tumor differentiation of patients
with LUAD. The deregulation of Linc00426 contributes to a notable impairment in proliferation, invasion, metastasis, and epithelial–mesenchymal transition (EMT) in vitro and in vivo.
Mechanistically, the deregulation of Linc00426 could reduce cytoskeleton rearrangement and matrix metalloproteinase expression. Meanwhile, decreasing the level of Linc00426 or increasing
miR-455-5p could down-regulate the level of UBE2V1. Thus, Linc00426 may act as a competing endogenous RNA (ceRNA) to abate miR-455-5p-dependent UBE2V1 reduction. We conclude that Linc00426
accelerates LUAD progression by acting as a molecular sponge to regulate miR-455-5p, and may be a potential novel tumor marker for LUAD. SIMILAR CONTENT BEING VIEWED BY OTHERS LNCRNA UCC
PROMOTES EPITHELIAL–MESENCHYMAL TRANSITION VIA THE MIR-143-3P/SOX5 AXIS IN NON-SMALL-CELL LUNG CANCER Article 06 April 2021 LINC00152 INDUCED BY TGF-Β PROMOTES METASTASIS VIA HUR IN LUNG
ADENOCARCINOMA Article Open access 07 September 2022 LNCRNA LINC00649 RECRUITS TAF15 AND ENHANCES MAPK6 EXPRESSION TO PROMOTE THE DEVELOPMENT OF LUNG SQUAMOUS CELL CARCINOMA VIA ACTIVATING
MAPK SIGNALING PATHWAY Article 28 February 2022 INTRODUCTION Lung cancer, with a 5-year overall survival rate of 15%, is a predominant cause of cancer-related deaths among males1.
Approximately 80% of lung cancers are non-small-cell lung cancers (NSCLCs)2. The two main types of NSCLCs are lung adenocarcinoma (LUAD) and lung squamous cell carcinoma3. Patients with LUAD
often die of relapse and metastasis due to the potential of progressing to hematogenous metastasis4. Despite the significant progress in the treatment of LUAD in recent years, its prognosis
remains poor, particularly for cases with lymph node metastasis or distant tumor metastasis. Thus, elucidating the potential mechanisms underlying the LUAD progression to identify novel
targets for effective intervention is urgently needed. Long non-coding RNAs (LncRNAs) are a class of non-protein-coding RNAs with over 200 nucleotides in length and can modulate gene
expression at transcriptional or post-transcriptional level5,6,7. LncRNAs function as oncogenes or tumor suppressors by activating or silencing the expression of protein-coding genes and
participating in cell proliferation, development, invasion, migration, and apoptosis8,9. Their expression is frequently dysregulated in multiple types of tumor such as lung cancer10. For
example, a novel LncRNA of LUADT1 promotes LUAD proliferation via epigenetic deletion of p2711. The enhanced expression of LncRNA of ZXF1 facilitates the invasion and metastasis of LUAD12.
Moreover, another lowly expressed LncRNA of RPLP0P2 is associated with poor prognosis and can abolish the cell abilities of proliferation and adhesion in LUAD13. Recently, a type of LncRNAs
has been classified as ceRNAs which are involved in the crosstalk among LncRNAs, mRNAs, and their shared microRNAs (miRNAs). This revealed a new regulatory mechanism by which LncRNAs and
mRNAs communicate with each other by competing for the common miRNAs response element. Here, we investigated the role of the Linc00426-miR-455-5p-UBE2V1 axis in regulating the process of
LUAD progression and the underlying mechanisms. Given its significance in the process of tumor progression, Linc00426 may act as a promising predictive biomarker for LUAD, and proper
regulation of the Linc00426-miR-455-5p-UBE2V1 axis may be a novel synergistic strategy for inhibiting the process of tumor progression. MATERIALS AND METHODS LNCRNAS MICROARRAY AND DATA
ANALYSES For LncRNAs microarray analyses, six LUAD tissue samples (three LUAD tissues with lymph node metastasis and three without) were obtained from the Affiliate Hospital of Weifang
Medical University. The levels of LncRNAs were profiled using Human LncRNA Array V3.0 (Arraystar, Rockville, Maryland, USA), and analyzed by KangChen Bio-tech Inc. (Shanghai, China). The
differentially expressed genes were employed for cluster analysis of samples and heat map generation. HUMAN TISSUE SPECIMENS This study was approved by the Ethics Committee of Weifang
Medical University and written informed consents were obtained from all subjects included in the research. The tissues were obtained from the Affiliate Hospital of Weifang Medical
University. Based on the literature review and our preliminary experiments, a significant level of α = 0.05, power of test = 0.9, and 144 specimens of clinical tissues (72 tissues with lymph
node metastasis and 72 without) were selected. Frozen tumor samples were used for total RNA extraction using the TRIzol method. All tumor tissues were confirmed by two experienced
pathologists. Clinicopathological characteristics of patients with LUAD were shown in Table 1. CELL CULTURE AND REAGENTS Cells (A549, H1975, H1299, BEAS-2B, and HEK-293T) were purchased from
the American Type Culture Collection. BEAS-2B and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), with H1299 and H1975 in RPMI-1640 medium and A549 in F12-K at 37
°C with 5% CO2. All cell media were supplemented with 10% fetal bovine serum (FBS). WESTERN BLOT AND IMMUNOFLUORESCENCE Total proteins were extracted from LUAD cell lines as previously
stated14. The following antibodies were used in this study: Vimentin (1:500), N-cadherin (1:500), E-cadherin (1:500), and UBE2V1 (1:500). The β-actin antibody was used as a control for
whole-cell lysates. Antibodies were purchased from Abcam (ab92547, ab18203, ab194982, ab151725, and ab181602). A549 cells were plated on glass slides (1 × 104 cells/well) and incubated for
approximately 24 h at 37 °C. The cells were then washed with PBS, and harvested, followed by being subjected to 4% paraformaldehyde for fixation, 0.1% Triton X-100 for permeabilization, and
serum working fluid for blocking nonspecific binding, successively. The cells were then incubated with fluorescence secondary antibodies and counterstained with 4′,6-diamidino-2-phenylindole
(DAPI) the next day. Confocal laser scanning microscope (SP8, Leica, Germany) was used to obtain the fluorescence images. CELL PROLIFERATION After being digested with trypsinase, cells were
suspended in complete medium. Three wells for each group were seeded at 5 × 104 cells/well and the cells were incubated at 37 °C with 5% CO2. From the second day after plating, the cell
counts were detected by Celigo Image Cytometer (Celigo, Nexcelom, USA) once a day for 5 days. At each time point, cell counts of the three wells in each group were presented as the mean
value. The cell growth curves of each group were plotted for five consecutive days. WOUND-HEALING ASSAYS For wound-healing assays, all cells were cultured in medium with low concentration
serum (supplemented with 0.5% FBS). A scratch tool was used to generate scratches gently in the lower central part of the 96-well plates. Serum-free culture medium was used to rinse the
cells gently, and culture medium with low concentration serum was added and the microscopic observation was performed at 0 h. Then, the Celigo Image Cytometer was used to analyze the images
after cell migration for 24 and 32 h in the same field and photographed, respectively. Celigo Image Cytometer was used to analyze the scanning images and obtain the cell migration area. CELL
INVASION The invasion of A549 cells was examined based on the number of cells through Matrigel-coated transwell inserts. In brief, 2 × 105 cells were seeded into 24-well plate-sized inserts
with Matrigel (BD Biosciences, USA). The cells were plated in serum-free medium, with the bottom chamber containing that with 10% FBS. After being incubated for both 24 and 32 h, cells
without penetrating the small holes were gently removed by cotton swabs. The cells were then fixed and stained with Giemsa. Cells from five random fields of view were counted under an
optical microscope (Olympus, Tokyo, Japan). RNA EXTRACTION AND REAL-TIME QUANTITATIVE PCR (RT-QPCR) RNA level was measured by RT-qPCR. Total RNA was extracted by TRIzol reagent according to
the manufacturer’s manual. M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) was used for LncRNA reverse transcription. cDNA was synthesized by a MMLV first-strand cDNA synthesis
kit (Promega, WI, USA). SYBR Green quantitative PCR kit (Bio-Rad, Hercules, CA, USA) was used to detect Linc00426 levels in tissues and cells. U6 and GAPDH were used as internal references
for normalizing the target genes. LUCIFERASE REPORTER ASSAY Linc00426 and UBE2V1 mRNA with mutant (Mut) or wild type (Wt) within miR-455-5p binding sites were cloned into upstream of the
luciferase reporter. The pGL-3 Basic Luciferase Vector (Promega, USA) lacking the Linc00426 or UBE2V1 transcript inserts served as the control. The renilla reporter pRL (Promega, WI, USA)
plasmid was used to normalize the transfection efficiency. Data were expressed as relative light units for luciferase normalized to renilla luciferase activity. Three independent experiments
were performed. VECTOR CONSTRUCTION AND LENTIVIRUS INFECTION Linc00426 lentivirus was constructed by GeneChem Company (Shanghai, China). Sequences are as following: Sh-NC:
TTCTCCGAACGTGTCACGT, sh-Linc00426-1#: AAGGATGGAAATACAGAACAA, sh-Linc00426-2#: TAGGTCATAATTGCTTAACTA, and sh-Linc00426-3#: CTTGTCCAATTTGTAGGGAAA. Lentivirus infection was performed according
to the manufacturer’s protocol. Cells were observed under fluorescence microscope at 72 h after infection. Cells stably expressing Linc00426 shRNA were established by lentiviral transduction
and puromycin selection. Stable cells were cultured for further tests. Transfection was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
Anti-miR-455-5p and anti-NC (7 μg) were dissolved in 2 mL MEM. RNA IMMUNOPRECIPITATION (RIP) ASSAY An EZMagna RIP kit (Millipore, Billerica, MA, USA) was used following the manufacturer’s
protocol. A549 cells were lysed in complete RIP lysis buffer, and the cell extract was incubated with magnetic beads conjugated with specific antibodies or control IgG (Millipore) for 6 h at
4 °C. The beads were washed and then incubated with Proteinase K to detach proteins. The purified RNA was used for RT-qPCR analysis. BIOINFORMATICS ANALYSIS Target genes for miR-455-5p were
identified by starbase. PPI network was built on STRING (https://string-db.org/), and cytoscape was used for seeking target genes (namely the hub gene) for miR-455-5p. The prognostic values
of candidate genes were analyzed using the Kaplan–Meier Plotter (http://kmplot.com/analysis/), an online database that could assess the effects of 1,715 samples derived from ten independent
datasets15. The correlation between mRNA expression of candidate genes and overall survival (OS) in patients with lung cancer was assessed. TUMOR XENOGRAFTS BALB/c nude mice (4–5 weeks, _n_
= 40; Charles River) were carried out with the approval of the Ethics Committee (Committee on the Use of Live Animals in Teaching and Research) of Weifang Medical University. For tumor
xenograft assay, A549 cells were stably transfected with sh-NC and sh-Linc00426. Subsequently, mice were randomly grouped (n = 10) and the cells were xenografted (1 × 107/100 μL) into mice
after being harvested. The weight of mice and tumor volume were measured every 3 days. Tumor volume was calculated according to the formula (_x_2 × _y_)/2 where _x_ < _y_ (_x_ = width;
_y_ = length). Thirty-five days later, the mice were killed and the tumors were stripped for further analyses. For tumor metastasis assay, mice were randomly grouped (_n_ = 10), and 1 × 106
A549 cells transfected with sh-NC and sh-Linc00426 were intravenously injected into BALB/c nude mice which were sacrificed after 1 month. Lungs were removed for examination. HE staining was
used to observe metastatic lesions. HEMATOXYLIN-EOSIN (HE) STAINING The lungs were collected, fixed in 4% paraformaldehyde for 24 h, dehydrated by alcohol, cleared by xylene, and imbedded in
paraffin, successively. 4 mm sections were prepared and stained with HE. After being dewaxed with xylene, the sections were subjected to absolute ethanol, 95% ethanol, 80% ethanol, 75%
ethanol, and distilled water, successively. The sections were then stained with hematoxylin for 1 min, washed under running water, washed by distilled water, embedded in 95% ethanol, and
stained with eosin for 30 s, successively. 95% ethanol and absolute ethanol were used for dehydration, with xylene for clearing and neutral balsam for mounting. The histological changes were
analyzed under an optical microscope (Olympus, Tokyo, Japan). STATISTICAL ANALYSES All experiments were performed at least three times independently. Data were presented as the mean ±
standard deviation (SD). Shapiro-Wilk test was used to analyze the normality test. All data were found in a normal distribution, and variance was similar among the groups statistically
compared. All statistical analyses were carried out using SPSS18.0 software (IBM, Chicago, IL, USA). Statistical evaluation was performed using Student’s _t_ test (two-tailed) between two
groups. One-way analysis of variance (ANOVA) was employed to compare the difference among groups. The relationship between Linc00426 and miR-455-5p was also evaluated by Spearman correlation
analysis. Statistics with _P_ < 0.05 were considered as statistically different. RESULTS DIFFERENTIAL EXPRESSION PATTERNS OF LNCRNAS IN LUAD TISSUES WITH AND WITHOUT LYMPH NODE
METASTASIS Previous microarray results showed that some LncRNAs had a higher expression in LUAD tissues with lymph node metastasis. The array data have been published16 and the gene
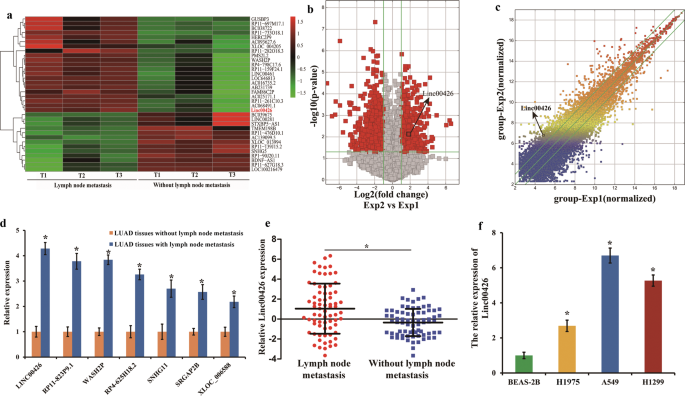
expression omnibus accession number is GSE115734. The microarray results showed higher expression of Linc00426 in LUAD tissues with lymphatic metastasis (Fig. 1a). The overview of scatter
and volcano plots displayed similar results (Fig. 1b and c). The classification could provide some clues for exploring the role of LncRNAs. To validate the microarray results, we selected
seven up-regulated LncRNAs with RNA lengths ranging from 1000 to 1100 nucleotides (Supplementary Table 1) for subsequent analyses. These LncRNAs belong to intergenic differential ones, and
their expression was analyzed using RT-qPCR. The results showed that the Linc00426, derived from the seven LncRNAs, presented the most significant different expression (Fig. 1d). The data
were consistent with the microarray results. We detected the expression of Linc00426 in 144 specimens of LUAD tissues by RT-qPCR, with 72 with lymph node metastasis and 72 without. The
results showed that the expression of Linc00426 was more prominent in LUAD tissues with lymph node metastasis compared with those without (Fig. 1e). Based on the LncRNA microarray data of
LUAD and bioinformatics algorithm, we focused on the biological function of Linc00426 and constructed a LncRNA–miRNA–mRNA network for the first time. UPREGULATION OF LINC00426 IN LUAD CELL
LINES AND TISSUES IS ASSOCIATED WITH THE CLINICOPATHOLOGICAL FEATURES OF PATIENTS WITH LUAD To investigate the role of Linc00426 in LUAD, we examined its expression in normal pulmonary
bronchial epithelial cells BEAS-2B and LUAD cells (H1975, A549, and H1299) by RT-qPCR. It was significantly up-regulated in all LUAD cells compared with that in normal pulmonary bronchial
epithelial cells BEAS-2B (Fig. 1f). Correlation of clinicopathological features and Linc00426 expression level was investigated among LUAD tissues. No significant correlation was found
between Linc00426 expression and age, gender of patients with LUAD. However, its expression was correlated with tumor size, tumor differentiation, lymph node metastasis, and distant
metastasis (Table 1). Hence, Linc00426 expression might be correlated with LUAD progression and play an important role in LUAD. These data suggest that Linc00426 could be a valuable tumor
marker for LUAD. DOWNREGULATION OF LINC00426 LEADS TO SIGNIFICANTLY INHIBIT PROLIFERATION, METASTASIS, AND INVASION OF LUAD IN VITRO The upregulation of Linc00426 in LUAD cell lines and
tissues implies that it may play a role of oncogenes in LUAD. We then examined its functional effects on LUAD cells. In this study, A549 cells were selected based on the higher expression of
Linc00426 than that in BEAS-2B. We constructed stable cell lines by using a lentivirus vector to intercede Linc00426 expression in A549 cells. All siRNAs significantly decreased the
expression of Linc00426. Knockdown efficiency was identified by RT-qPCR. The sh-Linc00426-1# displayed the highest efficiency, thus A549/sh-NC and A549/sh-Linc00426-1# (sh-Linc00426) were
used for further studies (Fig. 2a). To confirm the role of Linc00426 in the proliferation of A549, we performed the analysis on cell proliferation. Knockdown of Linc00426 suppressed the
proliferation of A549 cells compared with sh-NC (Fig. 2b). The wound-healing assay showed that Linc00426 knockdown suppressed the metastasis capacity of A549 cells compared with sh-NC (Fig.
2c). The transwell invasion assays revealed that Linc00426 knockdown inhibited the invasion of A549 cells (Fig. 2d). EMT is considered as an important mechanism involved in invasion and
metastasis. In this regard, we detected whether Linc00426 could mediate EMT in LUAD cells. Immunofluorescence and Western blot analyses revealed that Linc00426 knockdown suppressed the
expression of Vimentin and N-cadherin but elevated that of E-cadherin (Fig. 2e, f). These findings indicated that the downregulation of Linc00426 suppressed the proliferation, metastasis,
and invasion of LUAD cells in vitro. LINC00426 IS REQUIRED FOR CYTOSKELETON REARRANGEMENT AND MATRIX METALLOPROTEINASE EXPRESSION, AND DOWNREGULATION OF LINC00426 REPRESSES TUMOR GROWTH AND
METASTASIS OF LUAD IN VIVO The deregulation of matrix proteases, including matrix metalloproteinases (MMPs), is a common means for cancer to remodel the ECM, particularly in the initial
phases of tumor cell dissemination and cancer development progression17. Cytoskeleton rearrangement can promote the migration and invasion of non-small cell lung cancer cells18. Thus, we
detected the effects of Linc00426 on MMPs expression and cytoskeleton rearrangement in LUAD cells. MMP-2 and MMP-9 play an important role in tumor development progression by promoting
migration and invasion. The Western blot data revealed that the downregulation of Linc00426 reduced the levels of MMP-2 and MMP-9 in A549 cells (Fig. 3a). F-actin polymerization is essential
for the invasion of malignant tumor. We employed the analysis of F-actin polymerization to determine the effects of Linc00426 deletion on the invasion capacity of LUAD cells. Data
demonstrated polymerization and notable reduction for F-actin in A549/sh-NC and A549/sh-Linc00426 cells, respectively (Fig. 3b). Hence, Linc00426 played a vital part in cytoskeleton
rearrangement. LIMK and cofilin are two essential proteins involved in the regulation of F-actin polymerization19. Then we tested the phosphorylation level changes of LIMK and cofilin. Data
showed that the deletion of Linc00426 expression restrained the phosphorylation levels of LIMK and cofilin, and the cytoskeleton was eliminated (Fig. 3c). Overall, Linc00426 is required for
cytoskeleton rearrangement and MMPs expression. Based on the data from in vitro experiments, the effects of Linc00426 on LUAD tumorigenesis in vivo were investigated. A549/sh-Linc00426 or
A549/sh-NC was injected subcutaneously into BALB/c nude mice. The results showed that less A549 cells with Linc00426 knockdown developed into tumors than control vector cells did. Meanwhile,
the volume and weight of tumors derived from A549/sh-Linc00426 cells (_n_ = 10) were notably decreased compared with those from A549/sh-NC ones (_n_ = 10) (Fig. 3d). The abrogation of
Linc00426 was sufficient to impede orthotopic tumorigenesis of A549 cells in vivo (Fig. 3e). Then, we examined the metastasis behavior changes in primary tumor models after subcutaneous
injection of infective A549 cells. The number of metastatic nodules in the lung of nude mice with stable Linc00426 knockdown was substantially reduced compared with that in the control
vector group (Fig. 3f, g). Histopathological analysis by HE staining confirmed lung metastatic foci (Fig. 3h). The results suggest that the deregulation of Linc00426 represses the
tumorigenesis and metastasis of LUAD cells in vivo. LINC00426 ACTS AS A CERNA BY COMPETITIVE BINDING TO MIR-455-5P LncRNAs differ in subcellular distributions, with some predominantly in the
cytoplasm while others in the nucleus. Accumulating evidence shows that LncRNAs can interact with miRNAs competitively and regulate mRNA expression predominantly in the cytoplasm, thereby
affecting tumorigenesis and metastasis. We measured the location of Linc00426 through RT-qPCR based on the cytoplasm and nuclear RNA of A549 cells, respectively. The results showed that
Linc00426 was a cytoplasm-enriched LncRNA in A549 cells (Fig. 4a). Therefore, it might function as an endogenous molecular sponge to modulate miRNAs. To identify the putative functional
properties of Linc00426 in the tumorigenesis of LUAD and its connection with miRNAs expression, we analyzed interested miRNAs that putatively bind to Linc00426 using bioinformatics (AnnoLnc,
miRcode, and LncBase predicted v.2) tools. MiR-455-5p is considered to a consensus target gene (Fig. 4b). The GO analysis indicated that miR-455-5p affected a list of genes associated with
cell migration, cancer-related signal transduction (epidermal growth factor receptor and canonical Wnt signaling pathways), and cadherin binding was involved in cell-cell adhesion from three
aspects of biological process, cellular component, and molecular function (Fig. 4c). KEGG pathway analysis was performed to reveal the possible biological functions of miR-455-5p integrated
signature. The top KEGG pathways enriched for the miR-455-5p targets were mainly associated with cancer-relevant pathways and transcriptional misregulation in cancer (Fig. 4d). Such
pathways included chronic myeloid leukemia, Hippo, and TGF-beta ones. Subsequent bioinformatics analyses revealed a complementary sequence for miR-455-5p in Linc00426. Luciferase reporter
assays were conducted to illustrate the binding of miR-455-5p to Linc00426. The overexpression of miR-455-5p abolished the luciferase activity of the Wt reporter vector, exclusive of the
mutated reporter vectors (Fig. 4e). Further research using miR-455-5p was conducted. To identify the relationship between Linc00426 and miR-455-5p, we performed RIP assay. Levels of
Linc00426 and miR-455-5p were higher in the anti-Ago2 group than those in the anti-normal IgG group (Fig. 4f). Furthermore, we analyzed the levels of miR-455-5p in LUAD cells, the results
showed that the expression of miR-455-5p was significantly down-regulated in all LUAD cells compared with that in BEAS-2B (Fig. 4g). The effects of Linc00426 knockdown on miR-455-5p
expression were also examined in LUAD cells. MiR-455-5p was notably up-regulated after transfection of sh-Linc00426 into A549 cells (Fig. 4h). Our findings also suggested an inverse
correlation between Linc00426 and miR-455-5p (Fig. 4i). Linc00426 functions as an endogenous molecular sponge to down-regulate miR-455-5p by competitive binding to miR-455-5p. LINC00426 ACTS
AS A CERNA BY SPONGING MIR-455-5P AND MODULATING UBE2V1 It has been found that one of the molecular mechanisms by which LncRNAs regulate gene expression is to interact with miRNA as ceRNAs
that bind to miRNA response elements (MREs) and protect miRNAs from binding to and repressing target RNAs20. Using bioinformatics analysis (TargetScan) tools, we found that miR-455-5p was
putatively bound to 258 targeted genes. Then, we used PPI analysis and cytoHubba algorithm of cytoscape to detect the potential targets for miR-455-5p. And the results revealed 5 genes of
SMAD specific E3 ubiquitin protein ligase 2 (SMURF2), suppressor of cytokine signaling 3 (SOCS3), ubiquitin-conjugating enzyme E2 K (UBE2K), F-box, leucine rich repeat protein 7 (FBXL7) and
UBE2V1 as the hub ones (Fig. 5a, b). Then, the results of starbase showed that UBE2K and UBE2V1 had higher expression in LUAD tissues (Fig. 5c–g). Kaplan–Meier plots showed that higher
expression of UBE2V1 corresponded to a lower overall survival probability for LUAD patients. However, UBE2K had a higher overall survival probability (Fig. 5h, i). Therefore, we focused on
UBE2V1. To verify the potential interaction between miR-455-5p and UBE2V1 in LUAD cells, we cloned the wild-type or mutant UBE2V1 3′-UTR downstream of the luciferase reporter gene (Fig. 5j).
The plasmids were co-transfected with miR-455-5p into HEK-293T cells. The dual-luciferase reporter assay revealed that miR-455-5p was bound to UBE2V1 (Fig. 5k). We determined the expression
of UBE2V1 in BEAS-2B and LUAD cells (H1975, A549, and H1299) by Western blot analyses. The results showed that the expression of UBE2V1 was significantly up-regulated in all LUAD cells
compared with that in BEAS-2B cells (Fig. 5l). The invasion assay elucidated that UBE2V1 knockdown dramatically depressed cell invasion of A549 (Fig. 5m). These data suggest that UBE2V1 is
an oncogene in LUAD. Meanwhile, the Western blot results clarified that overexpression of miR-455-5p markedly reduced the protein levels of UBE2V1 (Fig. 5n). Previous studies reported that
LncRNAs can modulate many downstream genes to affect the biological functions of tumor cells21,22. We further investigated whether UBE2V1 is the downstream protein molecule for Linc00426. As
predicted, the knockdown of Linc00426 reduced the expression of UAB2V1 in A549 cells compared with sh-NC (Fig. 5o). MIR-455-5P REVERSES THE PROMOTING EFFECTS OF LINC00426 ON THE INVASION
AND EMT OF LUAD CELLS We further detected relevant regulatory relationship among Linc00426, miR-455-5p, and UBE2V1. The group of sh-NC + anti-miR-455-5p enhanced the UBE2V1 level, while,
sh-Linc00426+anti-NC significantly down-regulated the UBE2V1 level, which could be partially rescued by sh-Linc00426+anti-miR-455-5p (Fig. 6a). Figure 6b indicates that compared with sh-NC +
anti-NC, sh-Linc00426+anti-NC suppresses the expression of Vimentin and N-cadherin but elevates that of E-cadherin. Moreover, sh-NC + anti-miR-455-5p elevate the expression of Vimentin and
N-cadherin but suppress that of E-cadherin, which can be reversed by sh-Linc00426+anti-miR-455-5p. And down-regulated Linc00426 could suppress the EMT process stimulated by anti-miR-455-5p
in A549 cells. In addition, we found that anti-miR-455-5p promoted the invasion ability of A549 cells, which could be reduced by down-regulated Linc00426. And the pro-invasive effect was
contracted by co-transfected with anti-miR-455-5p (Fig. 6c). Collectively, these results suggest that Linc00426 accelerates LUAD progression by acting as a molecular sponge to regulate
miR-455-5p (Fig. 6d). DISCUSSION LUAD is the most malignant subtype of NSCLC. Its incidence and mortality have being increased significantly23. In recent years, the ectopic expression of
LncRNAs has drawn increased attention in cancer research since it plays an essential part in the proliferation, metastasis, and invasion of malignant tumors. Although numerous LncRNAs have
been found in LUAD, few further researches have been conducted. LncRNAs, transcripts with lengths ranging from 200 nt to 100 kb and widely distributed in the whole genome, do not encode
proteins but modulate the expression of coding genes or non-coding RNAs24,25. LncRNAs were initially regarded as ineffective junk products. At present, they are considered to participate in
vital regulatory functions, boosting yet another layer of complexity to our understanding of genomic modulation26. The diverse LncRNAs have been considered correlated with cell growth, cell
apoptosis and epigenetic modulation, transcription, and translation in a particular spatiotemporal manner27. They are transcribed by RNA polymerase II, with some of them further regulated
via splicing at the 5′ and 3′ ends and exported to the cytoplasm. Lnc-31 can control post-transcriptional gene expression which is required for myoblast proliferation and accelerate
Rho-associated c, oiled-coil containing protein kinase 1 (ROCK1) protein synthesis by stabilizing its translational activator, Y-box binding protein 1 (YB-1)28. LncRNAs located in the
cytoplasm could affect the mRNA level through ceRNAs. Diverse LncRNAs function as ceRNAs to adjust the EMT progression, such as LncRNA of HULC in hepatocellular carcinoma and LncRNA of H19
in colorectal cancer29,30. Linc00673 serves as a miR-150-5p sponge to adjust zinc finger E-box binding homeobox 1 (ZEB1) expression, thereby influencing the proliferation, migration,
invasion, and EMT of NSCLC31. Our study reported the upregulation of Linc00426 (ENST00000417079), a 1045 nt intergenic non-coding RNA located at chromosome13:30, 340, 270-30, 373, and 899,
in the majority of patients with LUAD. Its knockdown inhibits cell growth, invasion, metastasis, and F-actin expression, suggesting its vital role in the development and progression of LUAD.
With the development of bioinformatics technology, the genomics, transcriptome, and epigenomics of cancers were investigated32. It was reported that Linc00426 expression level is elevated
in doxorubicin resistant osteosarcoma and significantly correlated with unfavorable prognosis33. In the present study, we identified Linc00426 highly expressed in LUAD by microarray,
suggesting Linc00426 may function as an extensive tumor promoter. Therefore, elucidating its involvement in LUAD tumorigenesis needs further verification. LncRNAs and miRNAs often regulate
each other by a ceRNAs-related mechanism. In the present study, miR-455 could serve as an anti-oncogene in NSCLC through upregulation of ZEB1 and a potential therapeutic target for NSCLC34.
Lan et al35. found that Serine/threonine kinase 17b (STK17B) promotes the EMT process via activating AKT/GSK-3β/Snail signal pathway, and miR-455-3p has been identified as an upstream
regulator of STK17B. Circ0007142-miR-455-5p-SGK1 axis regulates cell proliferation, apoptosis, migration, and invasion of colorectal cancer cells36. It was reported that miR-455-5p
suppresses prostate cancer cellular proliferation and induces cell apoptosis by downregulating C-C Motif Chemokine Receptor 5 (CCR5)37. LncRNA of Gm4419 accelerates hepatic I/R injury by
targeting the miR-455 and SRY-Box transcription factor 6 (SOX6) axis38. Zhang et al.39 revealed a novel mechanism underlying MCM3AP-AS1 induced hepatocellular carcinoma metastasis by
regulating miR-455. Consequently, in our study, Linc00426 and UBE2V1 are significantly up-regulated when miR-455-5p is knocked down. Knockdown of Linc00426 inhibits cell migration, invasion,
and EMT. MiR-455-5p could reverse the biological function of Linc00426 in A549 cells. UBE2V1 encodes Ubiquitin Conjugating Enzyme E2V1, a ubiquitin-conjugating E2 enzyme variant (UEV)
protein. UEVs are similar in sequence to ubiquitin-conjugating E2 enzymes but lack their enzymatic activity40. UBE2V1 plays an important role in protein aggregate formation. Inhibition of
UBE2V1 decreases aggregate formation through enhanced ubiquitin proteasome system performance rather than autophagy, and may provide a novel therapeutic target for treating cardiac
proteinopathies41. UBE2V1 has been demonstrated as one of the key components of TRAF6 to control NF-κB activation42. Nevertheless, the role of UBE2V1 in cancers including LUAD and the
mechanisms involved are still largely unknown. Our study proved that Linc00426 could sponge miR-455-5p and adjust the expression of UBE2V1 indirectly. Indeed, there are some limitations in
the present work. First, the microarray analyses were performed in only two groups of samples, which limits further bioinformatics analyses and the search for other mechanisms of Linc00426
underlying LUAD progression. Second, A549 cells are good representative LUAD cells. Nonetheless, more representative cell lines should be employed to validate our results. Finally, the
animal experiment and the clinical study were performed to confirm the role of Linc00426 in LUAD. We did not study whether miR-455-5p could abolish the function of UBE2V1 in vivo. In future
studies, we will examine the effects of miR-455-5p inhibition in Linc00426 knockdown cells in vivo and investigate other mechanisms by which Linc00426 plays the role as an oncogene in LUAD.
Collectively, these results highlight the biomarker potential of Linc00426-miR-455-5p-UBE2V1 expression in LUAD, although it needs to be confirmed in a larger scale of independent samples.
To conclude, this research mainly focuses on the novel lncRNAs involving in proliferation, invasion, metastasis, and EMT of LUAD cells. We identified Linc00426 by screening the expression
profile of lncRNAs in LUAD tissues with or without lymph node metastasis. Then we discovered significantly up-regulated Linc00426 in LUAD tissues and cell lines. Animal experiments revealed
Linc00426 promotes tumorigenicity and metastasis in vivo. The cell biological experiments uncovered that Linc00426 serves as a ceRNA with miR-455-5p and regulates the expression of UBE2V1 in
LUAD cells. Thus, our findings suggest the novel lncRNA of Linc00426 as a tumor promoter and potential therapeutic biomarker for LUAD. AVAILABILITY OF DATA AND MATERIALS The dataset
supporting the conclusions of this article is included within the article. CHANGE HISTORY * _ 05 AUGUST 2024 A Correction to this paper has been published:
https://doi.org/10.1038/s41419-024-06952-8 _ REFERENCES * Torre, L. A. et al. Global cancer statistics, 2012. _CA Cancer J. Clin._ 65, 87–108 (2015). Article PubMed Google Scholar *
Goldstraw, P. et al. Non-small-cell lung cancer. _Lancet_ 378, 1727–1740 (2011). Article PubMed Google Scholar * Ettinger, D. S. et al. Non-small cell lung cancer. _J. Natl Compr. Canc
Netw._ 8, 740–801 (2010). Article CAS PubMed Google Scholar * Li, Y. et al. Transcriptome profiling of lncRNA and co-expression networks in esophageal squamous cell carcinoma by RNA
sequencing. _Tumour Biol._ 37, 13091–13100 (2016). Article CAS PubMed Google Scholar * Ponting, C. P., Oliver, P. L. & Reik, W. Evolution and functions of long noncoding RNAs. _Cell_
136, 629–641 (2009). Article CAS PubMed Google Scholar * Kretz, M. et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. _Nature_ 493, 231–235 (2013).
Article CAS PubMed Google Scholar * Wang, Y. et al. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. _Mol. Cell Biol._ 34, 3182–3193
(2014). Article PubMed PubMed Central Google Scholar * Wei, S. et al. Long noncoding RNAs in regulating adipogenesis: new RNAs shed lights on obesity. _Cell Mol. Life Sci._ 73,
2079–2087 (2016). Article CAS PubMed PubMed Central Google Scholar * Cech, T. R. & Steitz, J. A. The noncoding RNA revolution-trashing old rules to forge new ones. _Cell_ 157, 77–94
(2014). Article CAS PubMed Google Scholar * Naemura, M. et al. Long noncoding RNA ANRIL regulates proliferation of non-small cell lung cancer and cervical cancer cells. _Anticancer
Res._ 35, 5377–5382 (2015). CAS PubMed Google Scholar * Qiu, M. et al. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. _Cell
Death Dis._ 6, e1858 (2015). Article CAS PubMed PubMed Central Google Scholar * Zhang, L. et al. Enhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in
lung adenocarcinoma. _Biomed. Pharmacother._ 68, 401–407 (2014). Article CAS PubMed Google Scholar * Chen, J. et al. Low expression lncRNA RPLP0P2 is associated with poor prognosis and
decreased cell proliferation and adhesion ability in lung adenocarcinoma. _Oncol. Rep._ 36, 1665–1671 (2016). Article CAS PubMed Google Scholar * Wu, F. et al. The lncRNA ZEB2-AS1 is
upregulated in gastric cancer and affects cell proliferation and invasion via miR-143-5p/HIF-1alpha axis. _Onco Targets Ther._ 12, 657–667 (2019). Article CAS PubMed PubMed Central
Google Scholar * Gyorffy, B. et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. _PLoS ONE_ 8,
e82241 (2013). Article PubMed PubMed Central Google Scholar * Yang, Z. et al. Microarray expression profile of long non-coding RNAs in human lung adenocarcinoma. _Thorac. Cancer_ 9,
1312–1322 (2018). Article CAS PubMed PubMed Central Google Scholar * Chi, X. et al. lncRNA GAS5 promotes M1 macrophage polarization via miR-455-5p/SOCS3 pathway in childhood pneumonia.
_J. Cell Physiol._ 234, 13242–13251 (2018). * Li, H. et al. PTTG1 promotes migration and invasion of human non-small cell lung cancer cells and is modulated by miR-186. _Carcinogenesis_ 34,
2145–2155 (2013). Article CAS PubMed Google Scholar * Chen, P. et al. Expression levels and cotargets of miRNA1263p and miRNA1265p in lung adenocarcinoma tissues: Alphan exploration with
RTqPCR, microarray and bioinformatic analyses. _Oncol. Rep._ 41, 939–953 (2019). Article CAS PubMed Google Scholar * Cao, C. et al. The long non-coding RNA, SNHG6-003, functions as a
competing endogenous RNA to promote the progression of hepatocellular carcinoma. _Oncogene_ 36, 1112–1122 (2017). Article CAS PubMed Google Scholar * Liu, W. et al. Identification of
genes associated with cancer progression and prognosis in lung adenocarcinoma: analyses based on microarray from Oncomine and The Cancer Genome Atlas databases. _Mol. Genet Genom. Med._ 7,
e00528 (2019). Article Google Scholar * Hu, H. et al. Long non-coding RNA XLOC_000647 suppresses progression of pancreatic cancer and decreases epithelial-mesenchymal transition-induced
cell invasion by down-regulating NLRP3. _Mol. Cancer_ 17, 18 (2018). Article PubMed PubMed Central Google Scholar * Liu, B. et al. MiR-195 suppresses non-small cell lung cancer by
targeting CHEK1. _Oncotarget_ 6, 9445–9456 (2015). Article PubMed PubMed Central Google Scholar * Li, X. et al. Deregulated Gab2 phosphorylation mediates aberrant AKT and STAT3 signaling
upon PIK3R1 loss in ovarian cancer. _Nat. Commun._ 10, 716 (2019). Article PubMed PubMed Central Google Scholar * Iyer, M. K. et al. The landscape of long noncoding RNAs in the human
transcriptome. _Nat. Genet._ 47, 199–208 (2015). Article CAS PubMed PubMed Central Google Scholar * Yang, Y. et al. EGR1 interacts with DNMT3L to inhibit the transcription of miR-195
and plays an anti-apoptotic role in the development of gastric cancer. _J Cell Mol Med._ 23, 7372–7381 (2019). * Shi, X. et al. Long non-coding RNAs: a new frontier in the study of human
diseases. _Cancer Lett._ 339, 159–166 (2013). Article CAS PubMed Google Scholar * Dimartino, D. et al. The long non-coding RNA lnc-31 interacts with Rock1 mRNA and mediates Its
YB-1-dependent translation. _Cell Rep._ 23, 733–740 (2018). Article CAS PubMed PubMed Central Google Scholar * Li, S. P. et al. LncRNA HULC enhances epithelial-mesenchymal transition to
promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. _Oncotarget_ 7, 42431–42446 (2016). Article PubMed PubMed Central Google
Scholar * Liang, W. C. et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. _Oncotarget_ 6, 22513–22525 (2015). Article
PubMed PubMed Central Google Scholar * Lu, W. et al. Erratum to: Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial
mesenchymal transition by sponging miR-150-5p. _Mol. Cancer_ 16, 144 (2017). Article PubMed PubMed Central Google Scholar * Alvarez, M. J. et al. Functional characterization of somatic
mutations in cancer using network-based inference of protein activity. _Nat. Genet._ 48, 838–847 (2016). Article CAS PubMed PubMed Central Google Scholar * Wang, L. et al. Long
non-coding RNA LINC00426 contributes to doxorubicin resistance by sponging miR-4319 in osteosarcoma. _Biol Direct_ 15, 11 (2020). * Li, Y. J. et al. MicroRNA-455 suppresses non-small cell
lung cancer through targeting ZEB1. _Cell Biol. Int._ 40, 621–628 (2016). Article CAS PubMed Google Scholar * Lan, Y. et al. STK17B promotes carcinogenesis and metastasis via
AKT/GSK-3beta/Snail signaling in hepatocellular carcinoma. _Cell Death Dis._ 9, 236 (2018). Article PubMed PubMed Central Google Scholar * Wen, T. et al. Circular RNA circ_0007142
regulates cell proliferation, apoptosis, migration and invasion via miR-455-5p/SGK1 axis in colorectal cancer. _Anti-cancer Drugs_ https://doi.org/10.1097/CAD.0000000000000992 (2020). *
Xing, Q. et al. MiR-455-5p suppresses the progression of prostate cancer by targeting CCR5. _Biomed. Res. Int._ 2019, 6394784 (2019). Article PubMed PubMed Central Google Scholar * Ying,
D. et al. LncRNA Gm4419 induces cell apoptosis in hepatic ischemia-reperfusion injury via regulating the miR-455-SOX6 axis. _Biochem. Cell Biol._ 98, 474–483 (2020). Article CAS PubMed
Google Scholar * Zhang, H., Luo, C. & Zhang, G. LncRNA MCM3AP-AS1 regulates epidermal growth factor receptor and autophagy to promote hepatocellular carcinoma metastasis by interacting
with miR-455. _DNA Cell Biol._ 38, 857–864 (2019). Article CAS PubMed Google Scholar * Sancho, E. et al. Role of UEV-1, an inactive variant of the E2 ubiquitin-conjugating enzymes, in in
vitro differentiation and cell cycle behavior of HT-29-M6 intestinal mucosecretory cells. _Mol. Cell Biol._ 18, 576–589 (1998). Article CAS PubMed PubMed Central Google Scholar * Xu,
N. et al. Ube2v1 positively regulates protein aggregation by modulating ubiquitin proteasome system performance partially through K63 ubiquitination. _Circ. Res._ 126, 907–922 (2020).
Article CAS PubMed PubMed Central Google Scholar * Wu, Z. et al. Ubiquitin-conjugating enzyme complex Uev1A-Ubc13 promotes breast cancer metastasis through nuclear factor-small ka,
CyrillicB mediated matrix metalloproteinase-1 gene regulation. _Breast Cancer Res._ 16, R75 (2014). Article PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This
work was supported by National Natural Science Foundation of China (Nos. 81702932, 81872163, 81672631, 81402389, and 81641111), the Natural Science Foundation of Shandong Province (No.
ZR2019MH033). AUTHOR INFORMATION Author notes * These authors contributed equally: Chonggao Yin, Baogang Zhang AUTHORS AND AFFILIATIONS * Experimental Center for Medicine Research, Weifang
Medical University, 261053, Weifang, China Hongli Li * Department of Pathology, School of Clinical Medicine, Weifang Medical University, 261053, Weifang, China Hongli Li, Jun Bai, Liping
Zhang, Dandan Zhou, Quan Zheng & Baogang Zhang * School of Clinical Medicine, Weifang Medical University, 261053, Weifang, China Qingjie Mu, Lihong Shi & Wenxia Su * College of
Biological Science and Technology, Weifang Medical University, 261053, Weifang, China Guoxin Zhang * Department of Clinical Surgery, Affiliated Hospital of Weifang Medical University,
261053, Weifang, China Zhixin Shen * College of Nursing, Weifang Medical University, 261053, Weifang, China Yuanyuan Zhang & Chonggao Yin Authors * Hongli Li View author publications You
can also search for this author inPubMed Google Scholar * Qingjie Mu View author publications You can also search for this author inPubMed Google Scholar * Guoxin Zhang View author
publications You can also search for this author inPubMed Google Scholar * Zhixin Shen View author publications You can also search for this author inPubMed Google Scholar * Yuanyuan Zhang
View author publications You can also search for this author inPubMed Google Scholar * Jun Bai View author publications You can also search for this author inPubMed Google Scholar * Liping
Zhang View author publications You can also search for this author inPubMed Google Scholar * Dandan Zhou View author publications You can also search for this author inPubMed Google Scholar
* Quan Zheng View author publications You can also search for this author inPubMed Google Scholar * Lihong Shi View author publications You can also search for this author inPubMed Google
Scholar * Wenxia Su View author publications You can also search for this author inPubMed Google Scholar * Chonggao Yin View author publications You can also search for this author inPubMed
Google Scholar * Baogang Zhang View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Z.B. and Y.C. conceived and designed the experiments. S.Z.,
Z.Y., B.J., Z.L., Z.D., Z.Q., S.L., and S.W. performed the main experiments and analyzed the data. L.H., M.Q. and Z.G. wrote the manuscript. All authors read and approved the final
manuscript. CORRESPONDING AUTHORS Correspondence to Chonggao Yin or Baogang Zhang. ETHICS DECLARATIONS ETHICS APPROVAL All experiments were performed under the approval of the Ethics
Committee at the Weifang Medical University. CONFLICT OF INTEREST The authors declare that they have no conflict of interest. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. Edited by B. Rotblat SUPPLEMENTARY INFORMATION SUPPLEMENTARY MATERIALS RIGHTS AND PERMISSIONS
OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or
format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or
other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in
the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the
copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Li, H., Mu, Q., Zhang, G.
_et al._ Linc00426 accelerates lung adenocarcinoma progression by regulating miR-455-5p as a molecular sponge. _Cell Death Dis_ 11, 1051 (2020). https://doi.org/10.1038/s41419-020-03259-2
Download citation * Received: 03 May 2020 * Revised: 27 October 2020 * Accepted: 09 November 2020 * Published: 11 December 2020 * DOI: https://doi.org/10.1038/s41419-020-03259-2 SHARE THIS
ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard
Provided by the Springer Nature SharedIt content-sharing initiative