Lincrna 1700020i14rik alleviates cell proliferation and fibrosis in diabetic nephropathy via mir-34a-5p/sirt1/hif-1α signaling
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Long intergenic noncoding RNAs (lincRNAs) have been gradually identified to be functional in a variety of different mechanisms associating with development and epigenetic regulation
of cellular homeostasis. However, the study of lincRNAs in diabetic nephropathy (DN) is still in its infancy. Here, we have found dysexpressed long noncoding RNAs (lncRNAs) in renal tissues
of db/db DN mice compared with db/m mice by RNA sequencing. In this study, 5 lincRNAs were confirmed to express in a consistent trend among these DN-related lncRNAs both in vivo and in
vitro. Particularly, 1700020I14Rik was the downregulated one. Moreover, our data showed overexpression or knockdown of 1700020I14Rik could regulate cell proliferation and fibrosis in mouse
mesangial cells (MCs). Furthermore, 1700020I14Rik was found to interact with miR-34a-5p via both the directly targeting way by bioinformatic investigation and luciferase assay and the
Ago2-dependent manner by RIP assay. Results also displayed that overexpression of 1700020I14Rik inhibited cell proliferation and expressions of renal fibrosis markers through
miR-34a-5p/Sirt1/HIF-1α pathway in MCs under high glucose condition, while knockdown of 1700020I14Rik could increase cell proliferation and expressions of renal fibrosis markers. In
conclusion, these results provide new insights into the regulation between 1700020I14Rik and miR-34a-5p/Sirt1/HIF-1α signaling pathway during the progression of DN. SIMILAR CONTENT BEING
VIEWED BY OTHERS LONG NON-CODING RNA DLX6-AS1 IS THE KEY MEDIATOR OF GLOMERULAR PODOCYTE INJURY AND ALBUMINURIA IN DIABETIC NEPHROPATHY BY TARGETING THE MIR-346/GSK-3Β SIGNALING PATHWAY
Article Open access 28 February 2023 BIOINFORMATICS ANALYSIS OF MIRNAS GERMACRONE PROTECTION ON DIABETIC NEPHROPATHY Article Open access 30 December 2024 KNOCKDOWN OF LNCRNA MALAT1
ATTENUATES RENAL INTERSTITIAL FIBROSIS THROUGH MIR-124-3P/ITGB1 AXIS Article Open access 23 October 2023 INTRODUCTION Diabetic nephropathy (DN) as one of the major microvascular
complications of diabetes is the primary cause of end-stage renal disease. It is typically characterized by renal proliferation, thickening of basement membranes, interstitial fibrosis, and
massive accumulation of the extracellular matrix1. However, the detailed mechanism of DN is still unknown to date. Increasing evidence shows that silent information regulator T1 (Sirt1) is
involved in many important biological processes, including inflammation, renal interstitial fibrosis, autophagy under hypoxia condition, ageing, and oxidative stress2. In recent years,
researchers also have focused on the relationship between Sirt1 and DN. A drop of Sirt1 expression was found in DN and knockdown of Sirt1 expression could abolish the beneficial effect of an
active component against renal damage in DN3,4,5,6, while activating Sirt1 reversed the inflammation and fibrosis induced by high glucose in rat glomerular mesangial cells (MCs) and
silencing Sirt1 could promote fibrosis factors and inflammation factors in high glucose-cultured rat glomerular measangial cells through promoting its downstream molecule hypoxia-inducible
factor-1α (HIF-1α) expression7. However, the potential regulation mechanism in Sirt1/HIF-1α signaling in DN remains to be fully elucidated. Noncoding RNAs, arbitrarily separated into long
noncoding RNAs (lncRNAs) (>200 nucleotides) and small ncRNAs (≤200), are involved in multifarious physiological and pathological progress. In the past decades, small ncRNAs, such as
microRNAs (miRNAs), have been intensely investigated and found to regulate various disease progresses, some of which are further treated as therapeutic targets. However, the functional role
of lncRNAs remains little known. The lncRNAs, a large class of pervasive transcriptions in the genome with poor or no protein-coding ability, have been found to participate in multifarious
physiological and pathological progress8,9. Long intergenic noncoding RNAs (lincRNAs) have been gradually identified to be functional in a variety of different mechanisms associating with
development and epigenetic regulation of cellular homeostasis. Although, its molecular mechanisms are largely unknown, numerous lncRNAs are reported to be involved in various kinds of
diseases and oncogenesis, including DN10,11,12,13. By using RNA sequencing (RNA-seq), we found a number of lncRNAs differently expressing in mouse model of DN, in which one of the
dysexpressed lincRNAs-1700020I14Rik (ENSMUST00000147425) stood out and was chosen for further research. Previous researches have demonstrated that many lncRNAs function as competing
endogenous RNA (ceRNA) and competitively bind miRNAs to affect its function and prevented its targeted transcript from being degraded14,15,16. Therefore, we propose that 1700020I14Rik would
function as a ceRNA to regulate the miRNAs in DN. In the present study, the expression level of 1700020I14Rik was decreased in DN in vivo and in vitro as detected by real-time quantitative
PCR (qRT-PCR). In addition, overexpression of 1700020I14Rik inhibited cell proliferation and fibrosis in MCs, while 1700020I14Rik knockdown induced the proliferation and fibrosis of MCs with
low glucose. Moreover, 1700020I14Rik was confirmed to interact with miR-34a-5p by bioinformatic analysis, dual-luciferase reporter assay, and RIP. Furthermore, data showed that
1700020I14Rik increased Sirt1 expression via competitively binding miR-34a-5p. Additionally, 1700020I14Rik affected on cell proliferation and fibrosis of MCs by Sirt1/HIF-1α signaling
pathway. Taken together, our results indicate that 1700020I14Rik may play an important regulatory role in DN through miR-34a-5p/Sirt1/HIF-1α signaling. RESULTS LINCRNAS WERE DYSEXPRESSED IN
DN IN VIVO AND IN VITRO, INCLUDING 1700020I14RIK Our study showed 106 lncRNAs were abnormally expressed in renal tissues of db/db DN mice by RNA-seq using the following criteria: _p_-value
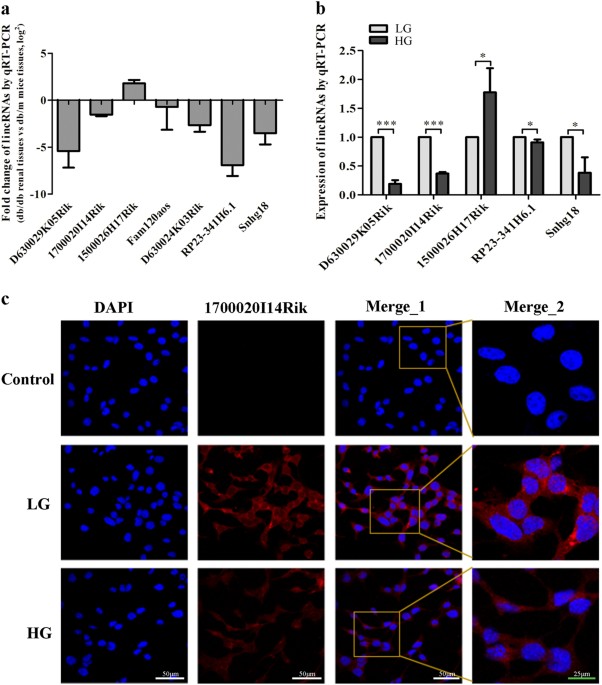
< 0.05 and _q_-value < 0.05 (Supplementary Table 1). On the basis of these data, we found there were 42 lincRNAs among these DN-related lncRNAs. Furthermore, the expressions of 7
lincRNAs with both high abundance in two groups (FPKM > 5) and fold changes >0.5 among them were verified in renal tissue of mice by qRT-PCR. Data showed that five lincRNAs showed an
expression of consistent with the result of RNA-seq, including four downexpressed and one upexpressed lincRNAs (Fig. 1a). Moreover, these five lincRNAs were verified in MCs by qRT-PCR and
data showed consistent results with those in renal tissues (Fig. 1b). Then, lincRNA 1700020I24Rik (ENSMUSG00000085438; Refseq NR_027832) was focused on for further study because it was not
only significantly downexpressed in DN in vivo and in vitro, but also displayed the highest conservative factor on the sequence comparing with their homologous sequence in human and high
abundance (FPKM > 11) among these five lincRNAs (Supplementary Table 2). Furthermore, FISH results confirmed that 1700020I24Rik was decreased in high glucose-cultured MCs compared with
that in MCs cultured with low glucose medium. Data also showed that since 1700020I24Rik located in both cytoplasm and nuclear in MCs, it was mainly distributed in the cytoplasm of cells
(Fig. 1c). Therefore, these demonstrate that 1700020I24Rik may be a DN-related factor. OVEREXPRESSION OF 1700020I14RIK INHIBITS PROLIFERATION AND FIBROSIS IN MCS UNDER HIGH GLUCOSE CONDITION
To study the effects of 1700020I14Rik in DN, 1700020I14Rik overexpression or knockdown was performed to investigate its functions. Firstly, the interfering efficiency of three siRNAs
against 1700020I14Rik was measured by qRT-PCR. Data suggested that siRNA-832 emerged the best knockdown efficiency, which was chosen for the following research. MCs transfected with
siRNA-832 or its matched control were named as 17Rik (−) or 17Rik (−) NC, respectively. Results also showed that MCs transfected with pcDNA3.1 (+)-1700020I14Rik or siRNA-832 could
effectively increase or decrease the expression of 1700020I14Rik (Fig. 2a). Moreover, CCK-8 assay showed the proliferation of MCs was decreased in 17Rik (+) group compared with that in 17Rik
(+) NC or HG control groups, while the cell proliferation was enhanced in 17Rik (−) group compared with that in 17Rik (−) NC or LG control groups (Fig. 2b). Furthermore, cell-cycle analysis
by flow cytometry showed that cells in the 17Rik (+) group were arrested in the S phase when compared with cells in the 17Rik (+) NC and HG control groups, whereas the proportion of cells
in S phase was observably increased in 17Rik (−) compared with 17Rik (−) NC or LG control groups (Fig. 2c). In addition, EdU assay showed that cell proliferation was significantly inhibited
in the 17Rik (+) group of MCs under high glucose condition compared with that in17Rik (+) NC and HG control groups, while 1700020I14Rik knockdown showed the opposite effect in the cells
cultured with low glucose (Fig. 2d). Consistent with these results, we observed decreased expression of the cell-cycle-related proteins cyclin D1, and increased expression of p21 in the
17Rik (+) group, while the expression of cyclin D1 and p21 was reversed respectively once the expression of 1700020I14Rik was knockdown in the 17Rik (−) group (Fig. 2e). Therefore, these
data indicate that overexpression of 1700020I14Rik inhibits cell proliferation in DN. To explore the effect of 1700020I14Rik on fibrosis in MCs, the expression levels of the well-known renal
fibrosis-related factors, collagen IV (Col-4), fibronectin (FN), and transcriptional regulatory factor-beta 1 (TGF-β1), were detected in this study. QRT-PCR data displayed that
overexpression of 1700020I14Rik significantly decreased the mRNA expressions of Col-4, FN, and TGF-β1, while 1700020I14Rik knockdown increased the expressions of Col-4, FN, and TGF-β1 (Fig.
3a–c). Meanwhile, western blot analysis showed the expressions of Col-4, FN, and TGF-β1 decreased in 17Rik (+) group compared with that in the 17Rik (+) NC and HG control groups (Fig. 3d).
However, the expressions of Col-4, FN, and TGF-β1 enhanced in 17Rik (−) group compared with that in 17Rik (−) NC or LG control groups (Fig. 3e). Furthermore, immunofluorescence assay
revealed that expressions of Col-4, FN, and TGF-β1 reduced in 17Rik (+) group compared with that in the 17Rik (+) NC and HG control groups (Fig. 3f). However, the expressions of Col-4, FN,
and TGF-β1 augmented in 17Rik (−) group compared with that in 17Rik (−) NC or LG control groups (Fig. 3g). Besides, ELISA assay also identified that overexpression of 1700020I14Rik
significantly inhibited the production and activation of TGF-β1, whereas, 1700020I14Rik knockdown increased its production and activation (Supplementary Fig 1). Overall, these data suggest
that 1700020I14Rik alleviates renal fibrosis in DN. 1700020I14RIK INTERACTS WITH MIR-34A-5P BY BOTH DIRECTLY TARGETING AND AGO2-DEPENDENT MANNERS To explore the mechanism of 1700020I14Rik in
proliferation and renal fibrosis of DN, the functions between lncRNAs and miRNAs are focused on because ceRNA mechanism plays an important role in diseases. Ten candidates of putative
targets for 1700020I14Rik were predicted by online databases (USCS, miRbase, and BiBiserv2 software) (Supplementary Table 3). Among these, miR-34a-5p was chosen for predicted candidate
because not only it was reported to be a DN-related miRNA17,18, but also it contained three binding sites in 1700020I14Rik transcript (Fig. 4a). Moreover, qRT-PCR data showed the expression
level of miR-34a-5p increased significantly in MCs cultured with high glucose, which displayed the opposite expression compared with 1700020I14Rik expression in DN (Fig. 4b). Furthermore,
data showed that overexpression of 1700020I14Rik reduced the expression of miR-34a-5p compared with that in the other two HG groups, whereas 1700020I14Rik knockdown elevated miR-34a-5p
expression in cells under low glucose condition (Fig. 4c). In addition, miR-34a-5p mimics or inhibitor was used to up or down the expression of miR-34a-5p in MCs cultured with high or low
glucose (Fig. 4d). Data also showed the overexpression of miR-34a-5p decreased the expression of 1700020I14Rik in cells with low glucose, while downexpression of miR-34a-5p increased the
expression of 1700020I14Rik in cells with high glucose (Fig. 4e). Therefore, the reciprocal regulation between 1700020I14Rik and miR-34a may play a role in DN. To determine whether
1700020I14Rik was directly repressed by miR-34a-5p, we used a luciferase reporter plasmid containing the 1700020I14Rik cDNA wild-type (1700020I14Rik wt) or seed-sequence mutant
(1700020I14Rik mut), then we performed dual-luciferase assays in HEK 293 cells. A GP-miRGLO-miR-34a-5p-inhibitor plasmid was also used in HEK 293 cells as positive control (PC). As expected,
the luciferase activity of 1700020I14Rik-wt was significantly decreased by miR-34a-5p mimics, while miR-34a-5p had no influence on 1700020I14Rik-Mut. And there was an apparent reduced
fluorescence activity caused in PC group by miR-34a-5p mimics (Fig. 4f). These results indicate that there is a direct interaction between 1700020I14Rik and miR-34a-5p. Additionally, miRNAs
have been found to bind their targets to depress and/or degrade RNA through RNA-induced silencing complex (RISC) in an Ago2-dependent manner19,20,21. Thus, RIP assay was performed using
anti-Ago2 in MCs cultured with high glucose to perform the relationship between 1700020I14Rik and Ago2. Results showed that both 1700020I14Rik and miR-34a-5p immunoprecipitated with Ago2
antibody were enhanced relative to IgG control (Fig. 4g). Taken together, these data confirm that 1700020I14Rik interacts with miR-34a-5p by both directly targeting way and Ago2-dependent
manner. MIR-34A-5P PROMOTES PROLIFERATION AND FIBROSIS IN MCS VIA SIRT1/ HIF-1Α SIGNAL PATHWAY To investigate the functions of miR-34a-5p in DN, we surveyed a panel of cell proliferation and
fibrosis of MCs transfected with miR-34a-5p mimics and inhibitor. The results of CCK-8 and EdU assays showed overexpression of miR-34a-5p increased cell proliferation, while inhibiting the
expression of miR-34a-5p reduced its proliferation (Fig. 5a, c). Moreover, the cell-cycle analysis results showed silencing of miR-34a-5p led to an improvement of cell proportion in S phase,
while overexpression of miR-34a-5p reversed the results (Fig. 5b). Furthermore, decrease-expression of miR-34a-5p reduced the expression of cyclin D1 and increased the protein level of p21,
however, the expression of cyclin D1 was increased and the expression of p21 was decreased when miR-34a-5p was overexpressed (Fig. 5d). Meanwhile, overexpression of miR-34a-5p enhanced the
expressions of Col-4, FN, and TGF-β1, however, inhibition of the miR-34a-5p downregulated their expressions (Fig. 5e). Therefore, the results indicate that miR-34a-5p promotes proliferation
and fibrosis of MCs by regulating cyclin D1 and p21. Furthermore, a highly conserved sequence was displayed that was complementary to the ‘seed sequence’ of miR-34a-5p and was identified
within the Sirt1 3′-UTR by bioinformatics analysis (Targetscan) (Fig. 6a). As known, Sirt1/HIF-1α signal pathway plays an important role in the proliferation and fibrosis of DN7.
Importantly, researchers found Sirt1 was a direct target for miR-34a-5p by luciferase assay12,22. In our study, we also found that miR-34a-5p could directly target to Sirt1 by luciferase
assay (Fig. 6b). Moreover, our qRT-PCR and western blot analysis results showed that overexpression of miR-34a-5p significantly downregulated the expression of Sirt1 but upregulated HIF-1α
in MCs with low glucose. However, the expressions of Sirt1 and HIF-1α were reversed when miR-34a-5p was silenced in MCs with high glucose (Fig. 6c–f). In addition, to further explore the
relationship between miR-34a-5p and Sirt1, siRNA against Sirt1 was used and co-transfected with miR-34a-5p inhibitor in cells cultured with low glucose. Results showed the downregulation
effect of miR-34a-5p inhibitor on HIF-1α was abolished by the intervention of si-Sirt1. Furthermore, western blotting showed that the effects of miR-34a-5p silence on increasing the
expressions of Col-4, FN, and TGF-β1 were impeded by si-Sirt1 (Fig. 6g). Thus, the data suggest that miR-34a-5p promotes fibrosis in DN might through directly regulating the expression of
Sirt1. 1700020I14RIK REDUCES THE EXPRESSIONS OF FIBROSIS FACTORS IN MCS BY REGULATING MIR-34A-5P/SIRT1/HIF-1Α SIGNALING To clarify whether the effect of 1700020I14Rik on renal fibrosis was
the interaction with miR-34a-5p through Sirt1/ HIF-1α pathway, we firstly identified the effect of 1700020I14Rik on the expressions of Sirt1 and HIF-1α. Results showed that overexpression of
1700020I14Rik increased the expression of Sirt1 and reduced HIF-1α, while knockdown of 1700020I14Rik decreased the expression of Sirt1 and upregulated HIF-1α (Fig. 7a, b). Furthermore,
western blotting showed that overexpression of 1700020I14Rik increased the expression of Sirt1 and reduced the expression of HIF-1α. However, these altered expressions of Sirt1 and HIF-1α
were blocked by overexpression of miR-34a-5p. Meanwhile, decreased Sirt1 and upregulated HIF-1α by knockdown of 1700020I14Rik was reversed in the presence of miR-34a-5p inhibitor (Fig. 7c,
d). Therefore, these results indicate that 1700020I14Rik regulates the expressions of Sirt1 and HIF-1α by regulating miR-34a-5p. Additionally, results showed that though the expressions of
renal fibrosis genes TGF-β1, FN, and Col-4 were reduced by overexpression of 1700020I14Rik, the expressions of these proteins were abolished by the participation of miR-34a-5p mimics.
Meanwhile, although the expressions of TGF-β1, FN, and Col-4 were upregulated by silence of 1700020I14Rik, these changes were blocked by miR-34a-5p inhibitor (Fig. 7e, f). Additionally, as
known, the fibrosis-related genes collagen prolyl 4-hydroxylases 1 (P4HA1), collagen prolyl 4-hydroxylases 2 (P4HA2), and procollagen-lysine (PLOD2) are HIF-1α direct targets genes23. Thus,
the effect of 1700020I14Rik on the expressions of P4ha1, P4ha2, and Plod2 was examined by real-time PCR in this study. Data showed that overexpression of 1700020I14Rik could decrease the
expression of P4ha1, P4ha2, and Plod2. However, knockdown of the expression of 1700020I14rik increased the expression of P4ha1, P4ha2, and Plod2 (Supplementary Fig 3). Furthermore, data
showed that the expression of 1700020I14Rik was decreased and the expression of miR-34a-5p was increased in the cells under high glucose, while the expressions of 1700020I14Rik and
miR-34a-5p were reversed when Sirt1 was silenced (Supplementary Fig 4). DISCUSSION In the present study, we observed that a novel lincRNA 1700020I14Rik suppressed the proliferation and
fibrosis of MCs cultured with high glucose medium through directly interacting with miR-34a-5p and antagonizes its function by regulating the expression of its target Sirt1 and downstream
proteins. LncRNA, initially thought to be a spurious transcriptional noise24, was hotly studied in recent years and found to participate in the gene expression, mammalian development, and
various kinds of diseases process8,25. According to their function roles in molecular mechanism, lncRNAs were divided into four subtypes: scaffolds, decors, guides, and signals9. Besides,
lncRNAs could also be classified into five categories according to the location with nearby protein-coding transcripts:26 sense and antisense lncRNAs—overlapping one or more transcripts on
the same or opposite strand, respectively; bidirectional lncRNAs—oriented head-to-head with a coding transcript within 1000 bp; intronic lncRNAs—overlapping the intron of one or more coding
transcripts; and lincRNAs—do not overlap coding transcripts. In this study, we paid attention on lincRNAs, as it is easier to interpret its gene expression patterns, sequence conservation,
and perturbation outcomes than those of transcripts from loci overlapping other gene classes27. On the basis of our RNA-seq data, 42 lincRNAs were dysexpressed in renal tissues of DN mice.
Moreover, seven lincRNAs were chosen for the candidates because their high abundance and significant different expression. Our results of qRT-PCR showed five lincRNAs were in the consistent
expression trend in vivo and in vitro. Interestingly, a highly abundant mammalian lincRNA, 1700020I14Rik was focused on because of the high conservative factor on the sequence comparing with
their homologous sequence in human. As a homologous gene of 1700020I14Rik, OIP5-AS1was reported to reduce cell proliferation by serving as a sponge or a ceRNA for HuR in human cervical
carcinoma HeLa cells28. Therefore, we speculated that 1700020I14Rik might also participate in cell proliferation and 1700020I14Rik might be an important factor in DN. 1700020I14Rik is a
lincRNA, which locates in chromosome 2 (Chr2: 119594296–119600744). However, there is no report about 1700020I14Rik in diseases up to now. Previous studies have identified lncRNAs function
mainly depending on their subcellular location. lncRNAs locating in cytoplasmic play a role in gene regulation by combining with specific proteins, and lncRNAs in nucleus mainly participated
in the transcription process and chromatin remodeling29,30. In this study, we found that 1700020I14Rik was a mainly cytoplasmic lincRNA in MCs and it was decreased in high glucose-cultured
MCs compared with that in MCs cultured with low glucose medium. High glucose condition is a key external stimulus that is extremely important for DN. Additionally, to investigate the
biological function of 1700020I14Rik in DN, we assessed the cell proliferation and the expressions of renal fibrosis factors in MCs under high or low glucose condition. Our studies revealed
that silencing of 1700020I14Rik promoted proliferation and fibrosis in MCs under low glucose condition, whereas overexpression of 1700020I14Rik expression led to inhibit proliferation and
fibrosis in MCs under high glucose condition. Combine with the results of downexpression of 1700020I14Rik in DN, it reveals that the downexpression of 1700020I14Rik represents a potential
risk for poor prognosis and contributes to DN development and progression. Accumulating evidence has demonstrated that lncRNAs could participate in multifarious diseases progress by acting
as ceRNAs to compete with miRNAs and regulate their functional role with their targets in a RISC manner. Lin et al. has demonstrated that lncRNA H19 could competitively bind miR-17-5p to
regulate YES1 expression and inhibit miR-17-5p-induced cell-cycle progression, while promote migration and invasion in thyroid cancer14. LncRNA HOTAIR, playing an oncogenic role in gastric
reported by Liu et al15, acted as a ceRNA that modulated the expression of human epithelial growth factor receptor 2 (HER 2) through working as a sink of miR-331-3p. Gas5 exerts
tumor-suppressive functions in human glioma cells by directly targeting miR-222 and regulating the expression of its targets BMF and Plexin C116. To explore the mechanism of 1700020I14Rik in
DN, we focused on its target miRNAs. By online databases (USCS, miRbase and BiBiserv2 software) prediction, ten candidates of putative targets for 1700020I14Rik were predicted and miR-34-5p
was chosen for further study for being a DN-related miRNA17,18 and containing three binding sites in 1700020I14Rik transcript. QRT-PCR data showed the expression level of miR-34a-5p
displayed the opposite expression compared with 1700020I14Rik expression and there was a reciprocal regulation between 1700020I14Rik and miR-34a in DN. Dual-luciferase reporter assay was
further used to explore the direct relationship between 1700020I14Rik and miR34a-5p. We identified that lincRNA 1700020I14Rik could directly bind to miR-34a-5p. Additionally, miRNAs have
been found to bind their targets to depress and/or degrade RNA through RISC in an Ago2-dependent manner. In the present study, by using RIP assay, we found that lincRNA 1700020I14Rik was in
a same RISC complex with miR-34a-5p. Collectively, the results display 1700020I14Rik interacts with miR-34-5p by both directly targeting and Ago2-dependent ways in DN. Furthermore, previous
microarray results have identified the overexpression of miR-34a in DN17. Moreover, downregulation of miR-34a could alleviate mesangial proliferation in vitro and glomerular hypertrophy in
early DN mice by targeting GAS118. In this study, we demonstrated that overexpression of miR-34a-5p improved the proliferation and fibrosis in mouse MCs, which played an inverse function
compared with 1700020I14Rik. Moreover, recent studies showed that miR-34a could inhibit SIRT1 expression by directly binding to the 3′ UTRs of its mRNAs and effectively decreased the protein
expression and inhibited the activity of SIRT121,31. Sirt1, a common member of the mammalian sirtuin family, functions as a NAD+-dependent protein deacetylase that has protective effects
against metabolic stress and plays a role in the progress of DN32,33. It was determined that Sirt1 inhibited FN and TGF-β1 expression in rat glomerular MCs (GMCs)21. Sirt1 deletion caused
renal injury in diabetes through increasing the expressions of endothelin-1 and TGF-β131. Reduction of Sirt1 by advanced glycation-end products (AGEs) increased FN and TGF-β1, two fibrotic
indicators in GMCs, and ultimately promoted the pathological progression of DN34. Moreover, HIF-1α, an oxygen-sensitive monitor and regulatory protein35, is a downstream molecule of Sirt1 in
hypoxia36. Previous study have revealed that silencing Sirt1 could promoted HIF-1α expression, but inhibited its downstream fibrosis factors and inflammation factors in high
glucose-cultured rat glomerular measangial cells under normal oxygen condition7. With these views, we confirmed that miR-34a-5p could directly target to Sirt1 by luciferase assay.
Furthermore, we conducted overexpression or knockdown of miR-34a, as we supposed, miR-34a-5p regulated the expression of Sirt1, HIF-1α and the downstream proteins. Moreover, there was no
effects of miR-34a-5p on MCs anymore when Sirt1 was knockdown, which suggested that miR-34a-5p might promotes proliferation and fibrosis in MCs culture in high glucose through Sirt1/HIF-1α
signaling pathway. On the other hand, we found that 1700020I14Rik could increase the expression level of Sirt1 but reduce HIF-1α expression, which showed an opposite regulation comparing
with miR-34a-5p in MCs. Besides, the expressions of fibrosis-related HIF-1α target genes-P4ha1, P4ha2, and Plod2 were regulated by1700020I14Rik. It indicated that 1700020I14Rik was relevant
with the fibrosis progress of DN. Furthermore, we performed the rescue experiment to find out the interaction between 1700020I14Rik and miR-34a-5p. These findings demonstrate that in high
glucose condition, though downregulated by high glucose medium, decreased 1700020I14Rik accelerated fibrosis by increased the expression of miR-34a-5p, meanwhile, the ability of competition
with the miR-34a-5p was gone down accordingly, and the expression of the miR-34a-5p target gene, Sirt1 was decreased, renal injury was further aggravated via Sirt1/HIF-1α signal mechanism in
MCs. In summary, our results indicated that high glucose medium could regulate lincRNA 1700020I14Rik and it was downexpressed in DN. Overexpression of 1700020I14Rik affected cell
proliferation and fibrosis though Sirt1/HIF-1α signal pathway. Moreover, the interaction between 1700020I14Rik and miR-34a-5p via directly targeting way and Ago2-dependent manner may be the
mechanism of the functions of 1700020I14Rik in DN. These data suggested a coherent mechanism for the role of 1700020I14Rik in cell proliferation and fibrosis during DN, and provided new
insights into the mechanisms of renoprotection associated with1700020I14Rik and miR-34a-5p/Sirt1/HIF-1α, which could lead to a novel therapeutic strategy for DN. MATERIALS AND METHODS
ANIMALS Eight-week-old male C57BL/KsJ background db/db mice and their age-matched heterozygote male db/m mice were purchased from NBRI (Nanjing, China). Mice with genetic defects in the
leptin receptor (db/db) have been widely used as type 2 diabetes model. Moreover, the db/db mouse has a long history as a model of human DN, and its key common features with the human
condition are renal hypertrophy, glomerular enlargement, albuminuria, and mesangial matrix expansion37. Several studies established that hyperglycemia was noted at the age of 4 week and
albumin excretion rates are higher by 8- to 62-fold in db/db mice beginning at the age of eight week38. Therefore, at eight weeks, db/db mice were regarded as early stage of DN mice39,40.
Then, the mice were divided into two groups: CON group (db/m, _n_ = 5) and DN group (db/db, _n_ = 5), and significant pathological characteristics were appeared (Supplementary Fig 2). All
procedures were in accordance with institutional guidelines for the care and use of laboratory animals at Chongqing Medical University. Approval of the Ethics Committee of Chongqing Medical
University was obtained for the study. CELL CULTURE The mouse MCs purchased from Academy of Sciences of Shanghai (Shanghai, China) were cultured in Dulbecco’s modified eagle medium (DMEM)
with 20% fetal bovine serum (FBS) at 37 °C in an atmosphere containing 5% CO2. According to the previous results41, MCs were stimulated with D-glucose at 5.5 mmol/L glucose plus 19.5 mmol/L
mannitol (low glucose group, LG) or at 25 mmol/L glucose (high glucose group, HG). High glucose stimulation had imitated the growth environment of MCs under the condition of DN, and low
glucose stimulation had imitated normal growth environment. BIOCHEMICAL AND HISTOLOGY ASSAYS Bayer glucometer was used to measure the blood glucose and 24 h urinary albumin was measured by
the coomassie brilliant blue (CBB) method. Renal tissue paraffin sections (4 µm thicknesses) subjected for hematoxylin-eosin (HE) staining were treated with hematoxylin and 1% ethanol eosin,
and then mounted by neutral gum for light microscopic observation. Five random images were chosen and analyzed with Image-Pro Plus (Media Cybernetics, Bethesda, MD, USA). RNA EXTRACTION,
LIBRARY CONSTRUCTION AND SEQUENCING Total RNA from three db/db mice and three db/m mice were isolated using TRIzol® Reagent (Life Technologies) in accordance with the manufacturer’s
instructions, as described previously42. RNA quality and quantity were measured using an Agilent 2100 Bioanalyzer (Agilent Technologies), and then these RNA samples were used for cDNA
library preparation and sequencing, which was conducted by Beijing Genomics Institute (BGI, Shenzhen, China). cDNA libraries were constructed using the TruSeq™ RNA Library Preparation Kit v2
protocol (Illumina), as described previously.44 cDNA libraries were sequenced using an Illumina HiSeq2000 sequencer (typical read lengths 90 bp) and 200 bp paired-end reads were generated.
RNA-SEQ READS MAPPING AND ANALYSIS The raw reads were firstly filtered by removing adapter sequences, low quality reads containing >50% bases with quality [QA] ≤ 15, and reads with >2%
undefined nucleotides [N]. Clean reads were aligned with Ensembl Mouse Genome using Tophat (v2.0.13). Transcripts were assembled and quantified using Cufflinks (v2.2.1). FPKM (fragments per
kilobase of transcript per million mapped reads) values were used to quantify the assembled transcripts. Differential expression was also tested using Cufflinks. Any transcript, with status
OK and _q_-value less than 0.05, is considered as differentially expressed between the groups. The biotypes of the differentially expressed transcripts were downloaded at Ensembl. PLASMID
TRANSFECTION The complementary sequence of 1700020I14Rik was PCR-amplified and ligated into pcDNA3.1 (+) vector (Invitrogen, US) to construct 1700020I14Rik (+) plasmid (clone loci: _BamH
I_/_EcoR V_). The primers used were 5′-TCCTGGTTCTTCCATCCTGT-3′ (forward) and 5′-ACGGCTTTCCTGTGTTGAGT-3′ (reverse). Transfections were performed using opti-MEM, lipofectamine 2000, and
lipofectamine 3000 reagents (Invitrogen, CA, USA) according to the manufacturer’s instructions. MiR-34a-5p mimics, inhibitors, and a matched miRNA negative control were synthesized and
purified by Shanghai GenePharma Co. (Shanghai, China). Three small interfering RNAs (siRNAs) (siRNA-969, siRNA-931, siRNA-832) duplexes against 1700020I14Rik and siRNA against Sirt1 were
synthesized by Shanghai Shenggong CO (Shanghai, China). MCs transfected with 1700020I14Rik (+) plasmid or pcDNA3.1 (+) plasmid were named as 17Rik (+) or 17Rik (+) NC, respectively. The
transfection efficacy was measured by qRT-PCR. The sequences of plasmids used in the manuscript were shown in Supplementary Table 4. REVERSE TRANSCRIPTION PCR (RT-PCR) AND QUANTITATIVE
REAL-TIME PCR (QRT-PCR) Total RNA was extracted from MCs using RNA TRIzol (Invitrogen, US) according to the manufacturer’s instructions. The PrimeScript RT reagent Kit (Takara, Dalian,
China) was used for cDNA synthesis. The quantification of miRNA, mRNA, or lncRNA was measured by qRT-PCR experiment with SYBR Premix ExTaqTM II (Takara, Dalian, China) using CFX96 PCR System
(Bio-Rad). Relative expressions were normalized to the expression of U6 or β-actin and calculated using 2−ΔΔCT method. All experimets were performed at least three times. Primers used for
PCR were listed in Supplementary Table 5. FLUORESCENCE IN SITE HYBRIDIZATION (FISH) For FISH analysis, the MCs were dividied into three goups, named as the control group, which was cultured
with low glucose medium and was not treated with fluorescence probe of 1700010I14Rik, the LG group, and the HG group, which were both treated with fluorescence probe but cultured with low
glucose medium and high glucose medium, respectively. Each group was repeated three times. The slides were fixed in 4% paraformaldehyde (Sigma) and before pre-hybridization, cells were
permeabilized with cold 0.1 % Triton X-100. Pre-hybridization buffer was discarded and hybridization was carried out with the 1700020I14Rik probe overnight. After washing with SCC buffer,
the coverslip was dyed with DAPI and fluorescence test was conducted with laser scanning confocal microscope. FLOW CYTOMETRIC ANALYSIS MCs transfected for 48 h in 6-well plates were
collected, washed with PBS for three times, and resuspended in 0.5 ml PBS, fixed in 3.5 ml 70% cold ethanol at 4 °C overnight. Then, the cells were centrifuged and washed for three times
with PBS, resuspended and stained in PI/Triton X-100 (20 µg PI/0.1% Triton X-100) including 0.2 mg RNase A at 37 °C. BD Flow cytometry was performed to analyze cell-cycle distribution by
standard procedure. CELL PROLIFERATION ASSAY Cell proliferation was assessed using Cell Counting Kit-8 (CCK-8) (Dingguo BioTechnology Co Ltd, Beijing, China) and a 5-ethynyl-2-deoxyuridine
(EdU) labeling/detection kit (RiboBio, Guangzhou, China). For CCK-8, MCs seeded in a 96-well plate at a density of 8 × 103 per well were transfected with plasmids, siRNA, mimics or inhibitor
for 48 h. Then, 10 µl CCK-8 regent was added to each well containing 100 µl DMEM medium and the plate was incubated for 2 h at 37 °C. A microplate reader (iMark; Bio-Rad) was used for
absorbance at 450 nm. For EdU assay, the MCs were cultured in 96-well plate and transfected with plasmids, siRNA, mimics or inhibitor for 48 h when cells at about 80% confluence. A volume of
50 µM EdU labeling medium was added into each well and cells were incubated for 2 h at 37 °C under 5% CO2. After fixed with 4% paraformaldehyde, the cells were treated with 0.5% Triton
X-100 and washed with PBS. Subsequently, anti-EdU working solution was used to stain the cells and 100 µl of Hoechst 33342 (5 µg/ml) was added to incubate cells at room temperature.
Fluorescent microscope (Olympus, Japan) was performed to observe EdU-positive cells. DUAL-LUCIFERASE REPORTER ASSAY The 1700020I14Rik cDNA cloned into a pmiR-RB-REPORT™ dual-luciferase
vector (RuiBio Corp., Guangzhou, China) to generate a pmiR-RB-REPORT™-1700020I14Rik wild-type, was named 1700020I14Rik wt. The mutant construct without predicting binding sequences of
miR-34a-5p was also cloned into pmiR-RB-REPORT™ vector and named 1700020I14Rik mut. The miR-34a-5p-inhibitor oligo was cloned in GP-miRGLO dual-luciferase vector (GenePharma Co., Shanghai,
China) to generate a GP-miRGLO-miR-34a-5p-inhibitor. All the recombinant plasmids were confirmed by restriction enzyme digestion and DNA sequencing. HEK 293 cells were transfected with
1700020I14Rik wt, 1700020I14Rik mut, miR-34a mimics, or mimics negative control (mimics NC) with lipofectamine 2000. HEK 293 cells were transfected with GP-miRGLO-miR-34a-5p-inhibitor,
miR-34a mimics, or mimics NC with lipofectamine 2000. After 48 h, the cells were conducted with Dual-Luciferase® Reporter Assay System (Promega, WI, USA). Firefly luciferase activity was
normalized to Renilla luciferase activity (seen detailed procedures for Supplemental Materials). Annealing method was conducted to generate a pair of most stable Sirt1 oligo, a pair of Sirt1
mutant oligo and miR-34a-5p-inhibitor sponge. All the sequence fragments were cloned in GP-miRGLO dual-luciferase vector (GenePharma Co., Shanghai, China), respectively, to generate a
GP-miRGLO-Sirt1 wild-type (Sirt1 wt), a GP-miRGLO-Sirt1 mutant (Sirt1 mut), and a GP-miRGLO-miR-34a-5p-inhibitor. All the recombinant plasmids were confirmed by restriction enzyme digestion
and DNA sequencing. HEK 293 cells were transfected with Sirt1 wt, Sirt1 mut, GP-miRGLO-miR-34a-5p-inhibitor, miR-34a mimics, or mimics NC with lipofectamine 2000. After 48 h, the cells were
conducted with Dual-Luciferase® Reporter Assay System (Promega, WI, USA). Firefly luciferase activity was normalized to renilla luciferase activity (seen detailed procedures for Supplemental
Materials). ELISA ANALYSIS Cell lysis buffer from each treatment group was collected and detected using a mouse TGF-β1 enzyme-linked immunosorbent assay (ELISA) Kit (Boster Bio-engineering
Co, Ltd, Wuhan, China), according to the manufacturer’s instruction (seen detailed procedures for Supplemental Materials). RNA IMMUNOPRECIPITATION (RIP) To determine whether 1700020I14Rik
was associated with the RNA-induced silencing complex (RISC), RIP assays were performed using the EZMagna RIP kit (Millipore, Billerica, MA, USA) and the Ago2 antibody (ab32381, Abcam,
Cambridge, MA, USA) following the manufacturer’s protocol. qRT-PCR analysis was performed to measure the expression levels of 1700020I14Rik and miR-34a-5p. Nomal mouse IgG (Millipore,
Billerica, MA, USA) was used as negative control (seen detailed procedures for Supplemental Materials). WESTERN BLOT ANALYSIS Total protein from MMCs was extracted by ice-cold RIPA lysis
buffer (Beyotime, Shanghai, China). Equal amount of protein samples were subjected into 10% SDS-PAGE (Beyotime, Shanghai, China), transferred onto 0.22 µm PVDF membranes (Millipore,
Temecula, CA), and blocked in 5% nonfat milk. Then the membranes were incubated with specific primary antibodies. The primary antibodies used in the study were the following: cyclin D1
(Abcam, USA, 1:10000), p21 (Boster, China, 1:400), Col-4 (Proteintech, USA, 1:1000), FN (Proteintech, USA, 1:1000), HIF-1α (Sangon,China, 1:500), Sirt1 (Sangon, China, 1:500), TGF-β1
(Proteintech, USA, 1:500), and GAPDH (Abcam, USA, 1:5000). ECL system (Millipore, Temecula, CA) was used to detect the immunoreactive bands and GAPDH antibody was used as control. Gray value
of protein bands were quantified by Image Lab software. IMMUNOFLUORESCENCE ASSAY For immunofluorescence analysis, the fixed cells were permeablized with 0.1% Triton X-100 and blocked in 5%
goat serum solution, then primary antibodies FN (Proteintech, USA, 1:50), Col-4 (Proteintech, USA, 1:50), or TGF-β1 (Proteintech, USA, 1:50) was added onto the coverslips at 4 °C for
overnight and Cy3-labeled secondary antibody (Proteintech, USA, 1:50) was treated onto coverslips for 1 h at room temperature. After treated with DAPI, coverslips were sealed with glycerine.
The images were observed with QImaging Micro-publisherTM 5.0 RTV (Olympus Corporation, Japan) and analyzed with Image-Pro Plus (Medium Cybemetics, Bethesda, MD, USA). STATISTICAL ANALYSIS
Statistical analysis was analyzed by SPSS19.0. All the results were presented as means ± SD. Student’s _t_-test (two groups) and one-way ANOVA (no less than three groups) with Tukey’s
Multiple Comparisons Test were used as appropriate to analyze the significance of differences between groups. A probability value _p_ < 0.05 was considered statistically significant.
REFERENCES * Yiu, W. H., Lin, M. & Tang, S. C. Toll-like receptor activation: from renal inflammation to fibrosis. _Kidney Int Suppl._ 4, 20–25 (2011). Article CAS Google Scholar *
Kume, S., Kitada, M., Kanasaki, K., Maegawa, H. & Koya, D. Anti-aging molecule, Sirt1: a novel therapeutic target for diabetic nephropathy. _Arch. PharmRes._ 36, 230–236 (2013). Article
CAS Google Scholar * Li, C. et al. Tetrahy droxystilbene glucoside ameliorates diabetic nephropathy in rats: involvement of SIRT1 and TGF-β1pathway. _Eur. J. Pharmacol._ 649, 382–389
(2010). Article PubMed CAS Google Scholar * Kitada, M. & Koya, D. SIRT1 in type 2 diabetes: mechanisms and therapeutic potential. _Diabetes Metab. J._ 37, 315–325 (2013). Article
PubMed PubMed Central Google Scholar * Chuang, P. Y. et al. Alteration of forkhead box O (foxo4) acetylation mediates apoptosis of podocytes in diabetes mellitus. _PLoS One_ 6, e23566
(2011). Article PubMed PubMed Central CAS Google Scholar * Shang, G. et al. 3,5-Diiodo-L-thyronine ameliorates diabetic nephropathy instreptozotocin-induced diabetic rats. _Biochim
Biophys. Acta_ 5, 674–684 (2013). Article CAS Google Scholar * Shao, Y., Lv, C., Wu, C., Zhou, Y. & Wang, Q. Mir-217 promotes inflammation and fibrosis in high glucose cultured rat
glomerular mesangial cells via Sirt1/HIF-1α signaling pathway. _Diabetes Metab. Res Rev._ 6, 534–543 (2016). Article CAS Google Scholar * Ponting, C. P., Oliver, P. L. & Reik, W.
Evolution and functions of long noncoding RNAs. _Cell_ 136, 629–641 (2009). Article PubMed CAS Google Scholar * Wang, K. C. & Chang, H. Y. Molecular mechanisms of long noncoding
RNAs. _Mol. Cell_ 43, 904–914 (2011). Article PubMed PubMed Central CAS Google Scholar * Spizzo, R., Almeida, M. I., Colombatti, A. & Calin, G. A. Long non-coding RNAs and cancer: a
new frontier of translational research? _Oncogene_ 31, 4577–4587 (2013). Article CAS Google Scholar * Wapinski, O. & Chang, H. Y. Long noncoding RNAs and human disease. _Trends Cell
Biol._ 21, 354–361 (2011). Article PubMed CAS Google Scholar * Wang, M., Wang, S., Yao, D., Yan, Q. & Lu, W. A novel long non-coding RNA CYP4B1-PS1-001 regulates proliferation and
fibrosis in diabetic nephropathy. _Mol. Cell Endocrinol._ 426, 136–145 (2016). Article PubMed CAS Google Scholar * Kato, M. et al. An endoplasmic reticulum stress-regulated lncRNA
hosting a microRNA megacluster induces early features of diabetic nephropathy. _Nat. Commun._ 7, 12864 (2016). Article PubMed PubMed Central CAS Google Scholar * Liu, L. et al. Long
noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. _FEBS J._ 12, 2326–2339 (2016). Article CAS Google Scholar * Liu, X. H. et al. LncRNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. _Mol. Cancer_ 13, 92 (2014). Article PubMed PubMed Central CAS Google
Scholar * Zhao, X. et al. Gas5 exerts tumor-suppressive functions in human glioma cells by targeting miR-222. _Mol. Ther._ 23, 1899–1911 (2015). Article PubMed PubMed Central CAS Google
Scholar * Chen, Y. Q. et al. Abated microRNA-195 expression protected mesangial cells from apoptosis in early diabetic renal injury in mice. _J. Nephrol._ 25, 566–576 (2012). Article
PubMed CAS Google Scholar * Zhang, L. et al. Down-regulation of miR-34a alleviates mesangial proliferation _in vitro_ and glomerular hypertrophy in early diabetic nephropathy mice by
targeting GAS1. _J. Diabetes Complicat._ 28, 259–264 (2014). Article PubMed PubMed Central Google Scholar * Yoon, J. H. et al. LincRNA-p21 suppresses target mRNA translation. _Mol.
Cell._ 47, 648–655 (2012). Article PubMed PubMed Central CAS Google Scholar * Chiyomaru, T. et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells.
_J. Biol. Chem._ 289, 12550–12565 (2014). Article PubMed PubMed Central CAS Google Scholar * Zhang, Z. et al. Negative regulation of lncRNA GAS5 by miR-21. _Cell Death Differ._ 20,
1558–1568 (2013). Article PubMed PubMed Central CAS Google Scholar * Lee, J. & Kemper, J. K. Controlling SIRT1 expression by microRNAs in health and metabolic disease. _Aging
(Albany NY)._ 2, 527–534 (2010). Article PubMed PubMed Central CAS Google Scholar * Zhu, S., Zhang, X. O. & Yang, L. Panning for long noncoding RNAs. _Biomolecules_ 3, 226–241
(2013). Article PubMed PubMed Central CAS Google Scholar * Zaratiegui, M., Irvine, D. V. & Martienssen, R. A. Noncoding RNAs and gene silencing. _Cell_ 128, 763–776 (2007). Article
PubMed CAS Google Scholar * Caley, D. P., Pink, R. C., Trujillano, D. & Carter, D. R. Long noncoding RNAs, chromatin, and development. _Sci. World J._ 10, 90–102 (2010). Article
CAS Google Scholar * Ulitsky, I. & Bartel, D. P. lincRNAs: genomics, evolution, and mechanisms. _Cell_ 154, 26–46 (2013). Article PubMed PubMed Central CAS Google Scholar * Kim,
J. et al. LncRNA OIP5-AS1_/_cyrano sponges RNA-binding protein HuR. _Nucleic Acids Res._ 18, 2378–2392 (2016). Article CAS Google Scholar * Tsai, M. C. et al. Long noncoding RNA as
modular scaffold of histone modification complexes. _Science_ 329, 689–693 (2010). Article PubMed PubMed Central CAS Google Scholar * Martianov, I., Ramadass, A., Serra, B. A., Chow, N.
SpringerAmpamp; Akoulitchev, A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. _Nature_ 445, 666–670 (2007). Article PubMed CAS Google
Scholar * Wang, G. et al. miR-34a-5p Inhibition alleviates intestinal ischemia/reperfusion-induced reactive oxygen species accumulation and apoptosis via activation of SIRT1 signaling.
_Antioxid. Redox Signal._ 24, 961–973 (2016). Article PubMed CAS Google Scholar * Huang, K. P. et al. AGEs-RAGE system down-regulates Sirt1 through the ubiquitin-proteasome pathway to
promote FN and TGF-β1 expression in male rat glomerular mesangial cells. _Endocrinology_ 156, 268–279 (2015). Article PubMed CAS Google Scholar * Bible, E. Diabetic nephropathy: Sirt1
attenuates diabetic albuminuria. _Nat. Rev. Nephrol._ 9, 696 (2013). Article PubMed Google Scholar * Mortuza, R., Feng, B. & Chakrabarti, S. SIRT1 reduction causes renal and retinal
injury in diabetes through endothelin 1 and transforming growth factor β1. _J. Cell Mol. Med._ 19, 1857–1867 (2015). Article PubMed PubMed Central CAS Google Scholar * Zbytek, B.,
Peacock, D. L., Seagroves, T. N. & Slominski, A. Putative role of HIF transcriptional activity in melanocytes and melanoma biology. _Dermatoendocrinol_ 5, 239–251 (2013). Article PubMed
PubMed Central CAS Google Scholar * Laemmle, A. et al. Inhibition of SIRT1 impairs the accumulation and transcriptional activity of HIF-1alpha protein under hypoxic conditions. _PLoS
One_ 7, e33433 (2012). Article PubMed PubMed Central CAS Google Scholar * Azushima, K., Gurley, S. B. & Coffman, T. M. Modelling diabetic nephropathy in mice. _Nat. Rev. Nephrol._
14, 48–56 (2018). Article PubMed CAS Google Scholar * Sharma, K., McCue, P. & Dunn, S. R. Diabetic kidney disease in the db/db mouse. _Am. J. Physiol. Ren. Physiol._ 284, F1138–F1144
(2003). Article CAS Google Scholar * Chen, H. et al. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice.
_Cell_ 84, 491–495 (1996). Article PubMed CAS Google Scholar * Makino, H. et al. Altered gene expression related to glomerulogenesis and podocyte structure in early diabetic nephropathy
of db/db mice and its restoration by pioglitazone. _Diabetes_ 55, 2747–2756 (2006). Article PubMed CAS Google Scholar * Sun, Y. et al. miR-451 suppresses the NF-kappaB-mediated
proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. _Mol. Cell Endocrinol._ 433, 75–86 (2016). Article PubMed CAS Google Scholar * Ji, H. et al. Deep
sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. _PLoS One_ 9,
e110314 (2014). Article PubMed PubMed Central CAS Google Scholar * Chen, M. et al. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from
the human colon cancer LIM1863 cell line. _Sci. Rep._ 6, 38397 (2016). Article PubMed PubMed Central CAS Google Scholar Download references ACKNOWLEDGEMENTS This study was supported by
the National Natural Science Foundations of China (81570747 and 81770811). AUTHOR INFORMATION Author notes * Co-first authors: Ailing Li and Rui Peng. * These authors Contributed equally
Ailing Li and Rui Peng. * Edited by G Calin AUTHORS AND AFFILIATIONS * Molecular Medicine and Cancer Research Center, Chongqing Medical University, 400016, Chongqing, China Ailing Li, Yan
Sun, Handeng Liu, Huimin Peng & Zheng Zhang * Department of Pathology, The People’s Hospital of Rongchang Distrct, 402460, Chongqing, China Ailing Li * Department of Bioinformatics,
Chongqing Medical University, 400016, Chongqing, China Rui Peng Authors * Ailing Li View author publications You can also search for this author inPubMed Google Scholar * Rui Peng View
author publications You can also search for this author inPubMed Google Scholar * Yan Sun View author publications You can also search for this author inPubMed Google Scholar * Handeng Liu
View author publications You can also search for this author inPubMed Google Scholar * Huimin Peng View author publications You can also search for this author inPubMed Google Scholar *
Zheng Zhang View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Zheng Zhang. ETHICS DECLARATIONS CONFLICT OF INTEREST
The authors declare that they have no conflict of interest. ADDITIONAL INFORMATION PUBLISHER'S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published
maps and institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY INFORMATION RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the
article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use
is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Li, A., Peng, R., Sun, Y. _et al._ LincRNA 1700020I14Rik alleviates cell
proliferation and fibrosis in diabetic nephropathy via miR-34a-5p/Sirt1/HIF-1α signaling. _Cell Death Dis_ 9, 461 (2018). https://doi.org/10.1038/s41419-018-0527-8 Download citation *
Received: 29 October 2017 * Revised: 11 February 2018 * Accepted: 09 March 2018 * Published: 27 April 2018 * DOI: https://doi.org/10.1038/s41419-018-0527-8 SHARE THIS ARTICLE Anyone you
share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the
Springer Nature SharedIt content-sharing initiative