Targeting N-glycosylation of 4F2hc mediated by glycosyltransferase B3GNT3 sensitizes ferroptosis of pancreatic ductal adenocarcinoma
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Pancreatic ductal adenocarcinoma (PDAC) remains a highly fatal malignancy partially due to the acquired alterations related to aberrant protein glycosylation that pathologically remodel
molecular biological processes and protect PDAC cells from death. Ferroptosis driven by lethal lipid peroxidation provides a targetable vulnerability for PDAC. However, the crosstalk between
glycosylation and ferroptosis remains unclear. Here, we identified 4F2hc, a subunit of the glutamate-cystine antiporter system Xc–, and its asparagine (N)-glycosylation is involved in PDAC
ferroptosis by N- and O-linked glycoproteomics. Knockdown of SLC3A2 (gene name of 4F2hc) or blocking the N-glycosylation of 4F2hc potentiates ferroptosis sensitization of PDAC cells by
impairing the activity of system Xc– manifested by a marked decrease in intracellular glutathione. Mechanistically, we found that the glycosyltransferase B3GNT3 catalyzes the glycosylation
of 4F2hc, stabilizes the 4F2hc protein, and enhances the interaction between 4F2hc and xCT. Knockout of B3GNT3 or deletion of enzymatically active B3GNT3 sensitizes PDAC cells to
ferroptosis. Reconstitution of 4F2hc-deficient cells with wildtype 4F2hc restores ferroptosis resistance while glycosylation-mutated 4F2hc does not. Additionally, upon combination with a
ferroptosis inducer, treatment with the classical N-glycosylation inhibitor tunicamycin (TM) markedly triggers the overactivation of lipid peroxidation and enhances the sensitivity of PDAC
cells to ferroptosis. Notably, we confirmed that genetic perturbation of SLC3A2 or combination treatment with TM significantly augments ferroptosis-induced inhibition of orthotopic PDAC.
Clinically, high expression of 4F2hc and B3GNT3 contributes to the progression and poor survival of PDAC patients. Collectively, our findings reveal a previously unappreciated function of
N-glycosylation of 4F2hc in ferroptosis and suggest that dual targeting the vulnerabilities of N-glycosylation and ferroptosis may be an innovative therapeutic strategy for PDAC.
Ferroptosis is a novel form of regulated cell death characterized by the iron-dependent unrestricted toxic accumulation of lipid peroxidation products and plasma membrane rupture and was
termed by Dixon et al. in 2012 [1]. Research in this field has revealed various targetable vulnerabilities in cell metabolism, redox homeostasis, and iron handling, which may provide
potential intervention targets for anticancer therapy [2, 3]. Among the numerous regulatory pathways involved in ferroptosis defense, the glutamate-cystine antiporter system Xc– is widely
known and consists of two subunits, namely, the heavy chain subunit 4F2hc (also known as CD98hc, encoded by the SLC3A2 gene) and the light chain subunit xCT (encoded by the SLC7A11 gene),
which mediate the synthesis of cysteine-derived antioxidants [4]. SLC7A11 has been identified as the target gene of nuclear factor erythroid 2-related factor 2 (NRF2), which is responsible
for maintaining redox homeostasis during oxidative stress [5, 6]. xCT predominantly functions in the intracellular transfer of extracellular cystine, and xCT seems to be more critical than
4F2hc in ferroptosis regulation. However, we noticed that 4F2hc, a glycoprotein, is extremely important for maintaining xCT protein stability and appropriate membrane localization. In terms
of structural biology, 4F2hc controls the intracellular trafficking and membrane topology of its heterodimerization partner [7] through polar and hydrophobic interactions. For example,
residues in 4F2hc form a short helix to fix xCT on the intracellular side [8]. Thus, 4F2hc is required for the recruitment of xCT to the plasma membrane, its degradation ultimately results
in xCT functional inactivation [9, 10]. In addition, recent studies have shown that deletion of 4F2hc causes the decompensation of system Xc–, indicating that 4F2hc plays an equally
important role in inhibiting ferroptosis [11,12,13]. However, as a membrane glycoprotein, 4F2hc has not been extensively studied in the context of ferroptosis.
Disease-associated stress may pathologically remodel the proteome or cause protein connectivity dysfunction, especially molecular chaperones [14]. Glycosylation, a widespread
posttranslational modification (PTM) [15], is a finely tuned enzymatic reaction process that occurs mainly in the endoplasmic reticulum (ER) and Golgi apparatus, where glycosyltransferases
and glycosidases add glycans to proteins and lipids. There are two types of glycosylation: N-glycosylation and O-glycosylation [16]. Glycosylation of proteins stabilizes proteins, helps
proteins move to the right location, and guides molecular chaperones to fold properly. Additionally, emerging evidence indicates that aberrant protein glycosylation plays a critical role in
cell death evasion, sustained proliferative signaling, and chemoresistance in various malignancies, including pancreatic ductal adenocarcinoma (PDAC) [17, 18]. Both glycosylation and
ferroptosis are physiological metabolic processes that may provide new therapeutic opportunities for cancer treatment. However, hitherto the crosstalk between glycosylation and ferroptosis
has not yet been largely revealed.
PDAC accounts for more than 90% of pancreatic cancer cases and is often life-threatening, with a 5-year relative survival rate of approximately 11%. According to its higher incidence and
mortality rates, PDAC is projected to rank as the second leading cause of cancer-related death by 2040 [19, 20]. Accumulating evidence indicates that targeting ferroptosis could exploit a
vulnerability in cancer, especially PDAC, which is closely intertwined with KRAS mutant-driven activation of the antioxidant system and is rich in iron [21]. Previous preclinical studies
have indicated that suppressing system Xc– in the extrinsic pathway (such as system Xc– inhibition and cysteine depletion [22]) or directly reducing the activity or expression of the core
antioxidant molecule in the intrinsic pathway (such as inhibitors targeted for GPX4 [23], FSP1 [24] and DHODH [25]) might effectively render PDAC cells susceptible to ferroptosis. But the
success of such therapeutic strategies remains limited because of ferroptosis resistance and some other unknown underlying mechanisms [26]. Therefore, a deeper understanding of ferroptosis
resistance may reveal new ferroptosis-related mechanisms and provide more optimized treatment options for PDAC.
In this study, we performed integrative N- and O-linked glycoproteomics and functional analyses to reveal a previously unrecognized coupling between PDAC ferroptosis and N-glycosylation of
4F2hc, and demonstrate that disturbs the process of 4F2hc glycosylation can induce susceptibility of PDAC to ferroptosis.
A total of 291 patients diagnosed with primary PDAC between January 2015 and July 2019 were consecutively recruited from the pathology archives of the Peking Union Medical College Hospital
(PUMCH) (Beijing, China). Patients who had received neoadjuvant therapy and who died owing to postoperative complications or lacked follow-up information were excluded from this
retrospective study. Hematoxylin and eosin (HE)-stained slides from all patients were retrieved and reviewed by two pathologists (ZL and SY) who were blinded to the patient’s clinical
outcomes. In cases of disagreement, a third pathologist (JC) confirmed the histological diagnosis. Detailed clinicopathological data and prognostic information of all patients were collected
from the medical records and telephone interviews. The TMA with a single 2-mm core per case was constructed using a Manual Tissue Microarrayer (MiniCore, Mitogen, Hertford, UK). Briefly,
representative tumor areas were marked on a HE stained slide and then punched from each donor formalin-fixed paraffin-embedded (FFPE) block for the recipient TMA blocks.
Human cell lines HPNE, PANC-1, MIA PaCa-2, BxPC-3, and AsPC-1 were obtained from the National Biomedical Cell Resource Center (Beijing, China) and identified by STR Profiling (D2081-2084) as
well as tested negative for contamination. Cells were maintained at 37 °C in 5% CO2 incubator. HPNE were grown in 75% Dulbecco’s modified Eagle’s medium (DMEM) without glucose (Sigma,
D5030-10 × 1 L; St. Louis, MO, USA) and 25% Medium M3 Base (Incell corp, M300F-500; Texas, USA), supplemented with 1% penicillin and streptomycin (Gibco, 1514022; Massachusetts, USA), 10%
fetal bovine serum (FBS) (Corning, 35-010-CV; New York, USA), 5.5 mM D-glucose (Sigma-Aldrich, G7021-100G), 10 ng/ml human recombinant EGF (PeProtech, AF-100-15; New Jersey, USA), 1 ×
GlutaMaxTM Supplement (Gibco, 35050061), and 750 ng/ml puromycin(Gibco, A1113803). The remaining cancer cells were maintained in DMEM (Corning, 10-013-CV) or RPMI 1640 (Corning, 10-040-CV)
supplemented with 10% FBS. All cell lines were cultured in 6–10 cm dishes or plated in 6–24 well plates for the indicated experiment. All chemical compounds used in cell culture experiments
included RSL3 (Selleck, S8155; Houston, USA), erastin (Selleck, S7242), imidazole ketone erastin (Selleck, S8877), Liproxstatin-1 (Selleck, S7699), ±-α-tocopherol (Selleck, S6104), UAMC-3203
(Selleck, S8792), Z-VAD-FMK (Selleck, S7023), Necrostatin-1 (Selleck, S8037), cycloheximide (Selleck, S7418), tunicamycin, and protein glycosylation inhibitor (Abcam, ab120296).
1 × 106 PANC-1 cells were plated in 75 cm2 cell culture flasks (Corning, 430640, New York, USA) until they reached 90% cell confluence and then incubated with the indicated concentration of
RSL3 or not for 12 h (n = 3 per group). Following treatment, cells were lysed using 300 μL 8 M urea with 1% protease inhibitor and centrifuged at 14,000 × g for 20 min at 4 °C, and the
supernatant was collected. The protein concentrations of the lysates were determined using the Bradford methods (Thermo Scientific, PierceTM BCA protein assay kit, 23225, Massachusetts,
USA). The details are provided in the “Supplementary Materials and Methods”.
To evaluate the stability of the 4F2hc protein (including but not limited to), cycloheximide (CHX) (20 μM) was added to indicate cells that had been treated with tunicamycin or transfected
with sgB3GNT3 for 0, 2, 4, and 8 h. Then collection each sample was collected and subjected to western blotting, and the relative intensities of 4F2hc protein were quantified using Image J
software.
To validate the glycosylation of 4F2hc proteins, PNGase F (New England BioLabs, P0704S, USA) and O-glycosidase (New England BioLabs, P0733, USA) were used according to the manufacturer’s
protocol. Briefly, 20 μg protein from the whole-cell lysates was added to a 10 μL total reaction volume comprising 1 μL of 10 × glycoprotein denaturing buffer and 9 μL water and then
subjected to denaturation by heating at 100 °C for 10 min. After cooling to room temperature, 2 μL of NP-40, 2 μL of 10 × GlycoBuffer 2, and water were combined to make up a 20 μL reaction
volume. Following incubation with or without 2 μL PNGase F at 37 °C overnight, the mixture was subjected to immunoblotting analysis with the 4F2hc antibody.
Total RNA was extracted from PANC-1 cells treated with or without RSL3 (n = 3 per group) using the TRIzol reagent (Invitrogen, 15596018, Carlsbad, CA, USA). The purity and concentration of
RNA were determined using a NanoDrop spectrophotometer (Thermo Scientific, Massachusetts, USA), and integrity was determined using an Agilent 2100 bioanalyzer (RNA 6000 Nano kit 5067-1511).
The details are provided in the “Supplementary Materials and Methods”.
FRGs were retrieved from the FerrDb database (http://www.zhounan.org/ferrdb) [27], which included 255 drivers, 208 suppressors, and 125 markers (Fig. S1A and Table S1). After removing
repetitive genes, 259 FRGs were eventually obtained and annotated into corresponding protein names for subsequent intersection with differential glycoprotein. A total of 207 known
glycosyltransferase genes were collected from the RNA-Seq data for TCGA pancreatic cancer subjects [28], which were used to identify hub gene sets by intersecting with differentially
expressed genes screened out from RNA sequencing for further analysis.
mIHC/IF staining was performed on TMA slides using an Opal Multiplex IHC Assay Kit (Akoya Biosciences, MA, USA), according to the manufacturer’s protocol. The details are provided in the
“Supplementary Materials and Methods”.
1 × 105 cells were plated in 6-well plates and incubated overnight, then treated with various compounds for 12 h. After treatment, for cell death analysis, viable cells and floating cells
were collected and stained with 5 μL FITC Annexin V and 5 μL propidium iodide (BD Bioscience, Apoptosis Detection Kit I, 556547) 15 min. For lipid ROS detection, 5 μM of C11-BODIPY 581/591
(Invitrogen, D3861) was added and incubated with cells for 30 min at 37 °C, 5% CO2 in an incubator. Cells for both cell death analysis and lipid ROS detection were harvested and resuspended
in 200 μL PBS, and then strained through a 40 μm cell strainer (Corning Falcon, 352350) for flow cytometric analysis (BD LSRFortessa). A minimum of 10,000 cells per well were analyzed in the
FITC channel or in combination with the PI channel. The FlowJo v10 software was used to analyze cell death and lipid peroxidation.
Female and male BALB/c nude mice (4 weeks old) were purchased from the VitalRiver Laboratory Animal Technology Co., Ltd. (Beijing, China), and housed in the Center for Experimental Animals
Research at the Institute of Basic Medical Science, Chinese Academy of Medical Science (CAMS). For the subcutaneous transplant tumor models, 2 × 106 of Cas9Control and B3GNT3KO PANC-1 cells
were suspended in 100 μL PBS and injected subcutaneously into the right side of immuno-deficient nude mice. Tumor growth was monitored using a Vernier caliper taken once per week for 28
days. For the orthotopic transplantation tumor models, 2 ×106 of Cas9Control and B3GNT3KO PANC-1 cells, and 2 ×106 of shControl and shSLC3A2 PANC-1 cells which stably expressed firefly
luciferase were resuspended in PBS separately and were carefully injected into the surgically exposed pancreatic tail on day 7, and the pancreatic peritoneal cavity abdominal wall and skin
were stitched. Tumor progression (shControl and shSLC3A2 groups) was monitored by bioluminescent imaging (IVIS Lumina III). To investigate the therapeutic effect of IKE (40 mg/kg) and its
combination with tunicamycin (1 mg/kg) in nude mice bearing shSLC3A2 PANC-1 cells xenografts, the mice were treated with the above drugs every other day. The mice’s survival and tumor growth
were also recorded. All tumors were surgically removed and processed for IHC on day 28. The tumor volume was calculated using the following formula: tumor volume = 1/2 × tumor length ×
tumor width2.
All statistical analyses were performed using the SPSS version. 22 or GraphPad Prism version 9. To compare two experimental conditions, the Mann-Whitney test or Student’s t-test was
performed for unpaired samples, and the Wilcoxon rank-sum test was applied for all paired t-test. A two-way analysis of variance (ANOVA) was performed to compare the multiple experimental
groups. Two-sided Pearson’s test or Spearman rank correlation test was used to identify correlations among the variables. The log-rank test and univariate and multivariate Cox regression
analysis were performed to estimate the survival and determine the independent prognostic factors, respectively. No statistical methods were used to predetermine the sample size. Each
experiment was repeated at least three times unless otherwise indicated in the figure legend. All data are presented as the mean ± SD unless otherwise stated. Statistical significance was
set at a threshold of P value less than 0.05.
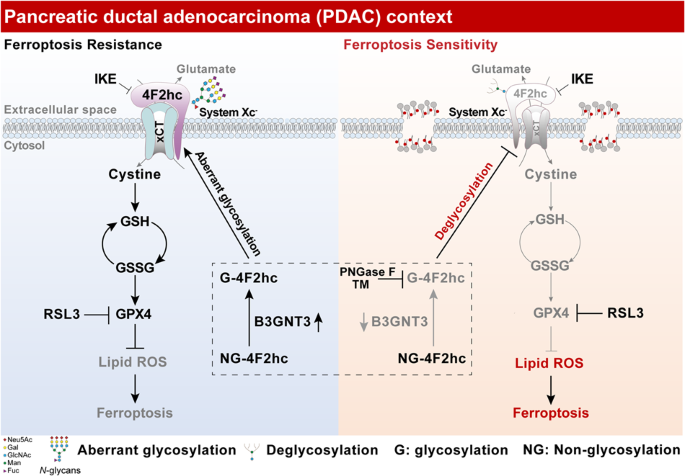
Aberrant glycosylation of the proteome protects pancreatic ductal adenocarcinoma (PDAC) from cell death induction [29]. However, its role in PDAC ferroptosis has not been well studied. To
this end, N-/O-glycoproteomics was performed to reveal the glycoproteomic characteristic of PDAC cells undergoing RSL3-induced ferroptosis (Fig. 1A). We first confirmed that most of the N-
and O-linked differentially expressed glycopeptides (DGPTs), including 530 (86.74%) N-DGPTs and 37 (6.06%) O-DGPTs, were upregulated in PANC-1 cells treated with RSL3 (Fig. 1B, S1B).
Accordantly, 191 (77.64%) N-linked differentially expressed glycoproteins (N-DGPs) and 21 (8.54%) O-linked DGPs were upregulated (Fig. S1C). Enrichment analyses of GeneOntology (GO) terms
indicated that those upregulated DGPs were enriched in transport, signal transduction, and cell death (Fig. S1D, F). Additionally, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analysis of the upregulated N-DGPs highlighted the pathway in cancer (Fig. S1E, G). Finally, seven candidate DGPs were identified including 4F2hc, GDF15, ANO6, TFR1, LAMP2, EGFR, and CD44
after the intersection of 211 N + O DGPs and 387 ferroptosis-related proteins (FRPs) (Fig. 1C, D).
A. Pipeline for identifying glycoproteins of interest shared by 211 DGPs and 387 ferroptosis-related proteins (FRPs). B Heatmap of normalized expression of differential glycopeptide across
the vehicle and RSL3 groups. The upper and bottom panels show the differential N- and O -glycopeptides, respectively. The dark blue- and red bars denote vehicle and RSL3, respectively. The
brown- and gray bars represent N-linked DGPs (N-DGPs) and O-linked DGPs (O-DGPs), respectively. C Venn diagram displaying the number of overlapping and unique proteins from 200 N-DGPs, 23
O-DGPs, and 387 FRPs. D Volcano plot showing 7 DGPs identified from N- and O-glycoproteomic. E LC–MS/MS-based analysis of the targeted differential N-linked glycosylation sites Asn365 of the
4F2hc in PANC-1 cells treated with or without 0.8 μM RSL3 for 12 h. The b11 and y4 ions represent the peptide fragmentation of Asn365 which was detected in RSL3-treated PANC-1 cells. F
Western blotting analysis of the glycosylation changes in 4F2hc protein extracted from PANC-1 cells was treated with PNGase F and O-glycosidase for 1 h at 37 °C. G PANC-1, AsPC-1, and MIA
PaCa-2 cells were treated with or without PNGase F for 1 h at 37 °C followed by immunoblot analysis. Purple- and cyan-triangle denote high- and low-glycosylation, respectively. H Cell
lysates from six PDAC tumors treated with or without PNGase F for 1 h at 37 °C. I Flow chart presenting an investigation to screen out the putative target genes from glycosyltransferase
genes (GTGs) and differentially expressed genes (DEGs) based on RNA-seq. J Left, Venn diagram displaying the number of overlapping genes from 207 GTGs and 5571 DEGs. Right, heatmap of all
the up- and down-regulated hub genes intersection from 207 GTGs and 5571 DEGs across the vehicle and RSL3 groups. The brown bar represents upregulated DEGs, and the gray bar represents
downregulated DEGs, the genes marked in red are four GTGs associated with PDAC prognosis. K qRT-PCR profiling of the expression of 4 selected upregulated genes involved in
glycosyltransferases in PANC-1 and MIA PaCa-2 cells treated with vehicle or RSL3. L Coomassie blue stained SDS gels of affinity-purified protein complexes co-immunoprecipitated by the 4F2hc
antibody from SLC3A2 overexpressed PANC-1 cells (line 3). Venn diagram displaying the 15 overlapping proteins from 1708 precipitated proteins and 185 glycosyltransferases (GTPs). M
Expression of 4F2hc, xCT (CST#12691, 35 kDa), B3GNT3, DHODH, GPX4, and FSP1 in PANC-1 cells treated with RSL3 at the range of concentrations for 12 h. N Western blot analyzing 4F2hc, xCT
(Abcam#175186, 55 kDa), B3GNT3, and GPX4 in human pancreatic epithelial nestin-expressing (HPNE) and PDAC cells (BxPC-3, MIA PaCa-2, PANC-1, AsPC-1). The western blot experiment was
representative of two biological replicates with similar results. All data are shown as the mean ± SD. Statistical significance among the indicated groups was assessed by ANOVA analysis or
unpaired Student’s t-test
Remarkably, 4F2hc has drawn our attention, as it is widely reported to be critical for ferroptosis defense but the role of 4F2hc glycosylation in ferroptosis is poorly understood. To
decipher the specific differential modification sites of 4F2hc affected by RSL3-induced ferroptosis, the tryptic glycopeptides of 4F2hc were analyzed by nanoscale LC–MS/MS. The results
showed that Asn365 was identified as a unique differential upregulated N-glycosylation site of 4F2hc (Fold change = 6.09; P-value = 0.02), and the intensity of Asn365-specific glycoforms
increased dynamically after ferroptosis activation (Fig. 1E). 4F2hc have four N-glycosylation sites include Asn365 predicted by NetNGlyc-1.0 (Fig. S1H). We confirmed that the N-glycosylation
of endogenous 4F2hc was completely inhibited and the molecular weight of 4F2hc was reduced from ~100 to ~70 kDa when PANC-1 cells lysates were treated with peptide-N-glycosidase F (PNGase
F), which is a recombinant glycosidase was used to remove asparagine-linked oligosaccharides from polypeptides, but not with recombinant O-glycosidase (Fig. 1F). Similar results were further
confirmed in PANC-1, AsPC-1, and MIA PaCa-2 cells and six human PDAC tissues when treated with PNGase F (Fig. 1G, H). Together, these results indicated that 4F2hc is a highly N-glycosylated
protein in PDAC and its N-glycosylation might play a potential role in ferroptosis execution.
To identify which glycosyltransferases are critical for glycosylation initiation during ferroptosis execution, 207 glycosyltransferase genes (GTGs) were collected from the reported
literature. Parallel RNA sequencing (RNA-seq) was performed in PANC-1 cells treated with RSL3 or not. We identified approximately 5571 differentially expressed genes (DEGs) (Fig. 1I). Then,
39 hub genes were identified after the intersection of the DEGs and GTGs, including 8 upregulated genes and 31 downregulated genes (Fig. 1I, J). KEGG enrichment analysis demonstrated that
ferroptosis was ranked among the top 10 pathways based on 2603 upregulated DEGs (Fig. S1I). Further analysis of 8 upregulated genes according to their mRNA expression from the TCGA datasets
revealed that just B3GNT3, B3GNT5, and ASGR1 were highly expressed in PDAC tissues compared to normal samples except for GCNT4 (Fig. S2A–D). High gene expression levels of B3GNT3 and B3GNT5
were associated with a poor prognosis for patients with PDAC, but ASGR1 and GCNT4 showed the opposite pattern (Fig. S2E–H). The qRT-PCR validation experiment showed that only B3GNT3 was
elevated considerably in indicated cells treated with RSL3 (Fig. 1K). Importantly, we identified B3GNT3, but not other glucosyltransferases, as a bona fide binding partner of 4F2hc protein
in SLC3A2OE PANC-1 cells analyzed by immunoprecipitation coupled with liquid chromatography-mass spectrometry/mass spectrometry (IP–LC–MS/MS) (Fig. 1L, Fig. S1J). Accordantly, 4F2hc was also
identified in B3GNT3OE PANC-1 cells (Fig. S1K). Co-immunoprecipitation results demonstrated that 4F2hc interacted with B3GNT3 in PANC-1 cells treated with or without RSL3 (Fig. S1L). We
further confirmed that the protein expression of 4F2hc and B3GNT3 gradually increased in PANC-1 cells treated with RSL3 in a dose-dependent escalation manner (Fig. 1M). In contrast, the
protein expression levels of xCT, DHODH, FSP1, and GPX4 showed the opposite trend (Fig. 1M). Finally, we confirmed that the protein levels of 4F2hc, B3GNT3, xCT, and GPX4 were highly
expressed in PDAC cells compared to normal human pancreatic epithelial nestin-expressing (HPNE) cells (Fig. 1N). These results imply that the glycosyltransferase B3GNT3 could be a potential
modulator in the response to ferroptosis, which might involve N-glycosylation of 4F2hc.
Inspired by these results, we sought to assess the clinical significance of 4F2hc and B3GNT3 in PDAC. We first confirmed that the protein levels of 4F2hc and B3GNT3 were highly expressed in
six paired PDAC tissues compared to adjacent normal tissues (Fig. 2A, B), which is consistent with the gene expression analysis from TCGA (Fig. S2I, J). We further confirmed that 4F2hc and
B3GNT3 were highly expressed (score of 2-3) in 59.92% (145/242) and 79.30% (203/256) of the 291 PDAC patient specimens, respectively (Fig. S2K, L). High expression of B3GNT3 positively
correlated with 4F2hc in patients with PDAC (Fig. 2C). 4F2hc exhibited a membranous staining pattern, whereas B3GNT3 staining was visualized as tan granules scattered around the cytoplasm
(Fig. 2D). In addition, the correlation analysis showed that high expression of 4F2hc was more frequent in PDAC with poor differentiation, and positive protein expression of B3GNT3 was more
significantly associated with American Joint Committee on Cancer (AJCC) III-IV stage and tumor stage in T1-2 (Fig. 2E, F) (Table S3).
A. 4F2hc and B3GNT3 protein expression in paired tumor and adjacent normal tissues from 6 PDAC patients was determined by western blotting. B Paired comparison of 4F2hc and B3GNT3 protein
expression. Statistical significance was determined by a two-tailed paired t-test. C Scatter plots showed the Pearson correlation of IHC scores of 4F2hc and B3GNT3. The purple line was a
linear fit changed with the expression of 4F2hc and B3GNT3. D Representative HE (top) and IHC (bottom) stained sections of 4F2hc and B3GNT3 in PDAC (scale bar = 100 μm). Scores 0, 1, 2, and
3 represent negative, low, moderate, and high, respectively. E, F Percentage of patient samples for 4F2hc and B3GNT3 in tumor differentiation and AJCC stage. The correlation between the
indicated molecule and the indicated clinicopathological characteristics was analyzed by the chi-square test. G, I, K Kaplan–Meier curves of progression‐free survival (PFS) stratified by
4F2hc (G) and B3GNT3 (I) and combined expression (K). H, J, L Kaplan–Meier curves of disease-specific survival (DSS) stratified by 4F2hc (H) and B3GNT3 (J) and combined expression (L). M
Representative multiplex immunohistochemistry images of the spatial distribution and abundance for 4F2hc, B3GNT3, xCT, NRF2, CK, and DAPI in TMAs sections from case 242 of PDAC. Glandular
intraepithelial neoplasia, (GIN). Scale bar: 200 µm. Statistical significance among the indicated groups was assessed by a one-tailed t-test, one-sided log-rank test, or Tarone-Ware test. *P