Mitochondrial RNA methyltransferase TRMT61B is a new, potential biomarker and therapeutic target for highly aneuploid cancers
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Despite being frequently observed in cancer cells, chromosomal instability (CIN) and its immediate consequence, aneuploidy, trigger adverse effects on cellular homeostasis that need to be
overcome by anti-stress mechanisms. As such, these safeguard responses represent a tumor-specific Achilles heel, since CIN and aneuploidy are rarely observed in normal cells. Recent data
have revealed that epitranscriptomic marks catalyzed by RNA-modifying enzymes change under various stress insults. However, whether aneuploidy is associated with such RNA modifying pathways
remains to be determined. Through an in silico search for aneuploidy biomarkers in cancer cells, we found TRMT61B, a mitochondrial RNA methyltransferase enzyme, to be associated with high
levels of aneuploidy. Accordingly, TRMT61B protein levels are increased in tumor cell lines with an imbalanced karyotype as well as in different tumor types when compared to control tissues.
Interestingly, while TRMT61B depletion induces senescence in melanoma cell lines with low levels of aneuploidy, it leads to apoptosis in cells with high levels. The therapeutic potential of
these results was further validated by targeting TRMT61B in transwell and xenografts assays. We show that TRM61B depletion reduces the expression of several mitochondrial encoded proteins
and limits mitochondrial function. Taken together, these results identify a new biomarker of aneuploidy in cancer cells that could potentially be used to selectively target highly aneuploid
tumors.
Aneuploidy, the immediate consequence of chromosomal instability (CIN) that describes an imbalanced karyotype, is frequently detected in solid and hematopoietic tumors [1, 2]. Accumulated
evidence indicates that particular aneuploidy conditions drive carcinogenesis [3]. Significantly, the genetic diversity associated with CIN and aneuploidy might lie behind the development of
drug resistance, high metastatic incidence and low survival rates in many cancer patients [4,5,6,7,8]. Paradoxically, aneuploidy is not only associated with cancer promotion but also with
the suppression of malignant growth [9]. In the short term, karyotypic changes normally results in a poor cellular fitness due to different stress factors, such as proteoxic and genotoxic
burden, overloaded endoplasmic reticulum, augmented glycolytic flux and mitochondrial activity, and an increase in reactive oxygen species (ROS) [10,11,12,13,14]. However, cancer cells are
generally characterized by an aneuploid and proliferative phenotype, which suggests that specific timely gains or losses of chromosomes should ultimately promote adaptation once the initial
challenge of gene imbalance has been overcome. In fact, different studies have shown that aneuploidy could facilitate the selection of cells with specific karyotypic compositions that allow
them to tolerate high stress conditions and adapt to an ever-changing environment [15,16,17,18,19]. These aneuploidy-induced responses include upregulation of autophagy, DNA repair
mechanisms, and antioxidant levels as well as more efficient bioenergetic and biosynthetic processes [13, 20,21,22,23]. Importantly, aneuploidy might make cancer cells more sensitive to
certain antitumoral therapies that exploit some of these aneuploidy coping mechanisms [24, 25]. Despite this knowledge, the mechanism whereby aneuploidy overcomes deleterious effects and
promotes cell selection is not fully understood [9].
Over 160 RNA modifications involving post-transcriptional changes in the chemical composition of different classes of nuclear and mitochondrial RNAs have been described to date, with tRNAs
and rRNAs containing the most numerous and chemically diverse modified ribonucleosides [26, 27]. These modifications range from the addition of a single methyl group in a nitrogenous base or
sugar residue to more complex molecular transformations. In this way, RNA function can be fine-tuned at different levels depending on the type, location and target of the modification
(reviewed in [28, 29]). Thus, certain RNA modifications are required for proper processing, folding, and stability during RNA biogenesis, while others play a crucial role in RNA-driven
protein synthesis by modulating translation efficiency and fidelity [30,31,32,33]. Many of these RNA composition changes are neither static nor stable [34, 35]; rather, they are dynamic
marks introduced and removed in a reversible process catalyzed by both epitranscriptomic “writer” and “eraser” enzymes, respectively, which can be potentially modulated in response to
different cellular stress cues [34, 35]. However, this stress-induced reprogramming of RNA modifications and, specifically, its likely association with aneuploidy and therapeutic utility
have yet to be explored.
In an in silico bioinformatics study, we identified the RNA methyltransferase enzyme TRNA Methyltransferase 61B (TRMT61B) as a potential aneuploidy biomarker. In this study, we examine in
detail the connection between this molecule and highly aneuploid cancers and explore its potential therapeutic value. Although it is encoded by the nuclear genome, TRMT61B is predominantly
localized in the mitochondrial compartment, where it catalyzes methylation at the N1 position of specific adenosine residues (referred to as m1A) present in the 3 major types of
mitochondrial RNA (mt-RNA) [36,37,38]. Although the molecular consequences of TRMT61B-mediated methylation on RNA biology have received some attention, its biological function is still
poorly understood [36,37,38]. Very recent studies suggest that this enzyme is involved in protein synthesis regulation, which is consistent with the role of TRMT61B mtRNA targets as critical
players in the mitochondrial translation mechanism [36,37,38]. However, a large body of research is required to address this issue in depth.
Our in silico and in vitro assays reveal a strong direct correlation between the degree of aneuploidy and TRMT61B protein levels in both NCI-60 cancer cell line collection and a more
specific panel of human melanoma cell lines. Furthermore, TRMT61B is overexpressed in different tumor types compared to normal tissues, and is positively associated with aneuploidy in
certain human cancers. We also demonstrate that cancer cells with high levels of aneuploidy are addicted to TRMT61B, since its elimination, using shRNA or CRISPR technology, has an
antiproliferative effect and is detrimental to mitochondrial function. Finally, xenotransplant experiments performed in both immunodeficient mice and zebrafish embryos show that TRMT61B
deficient cells with high aneuploidy levels are less effective in tumor formation. These findings heighten interest in this enzyme as a potential cancer target.
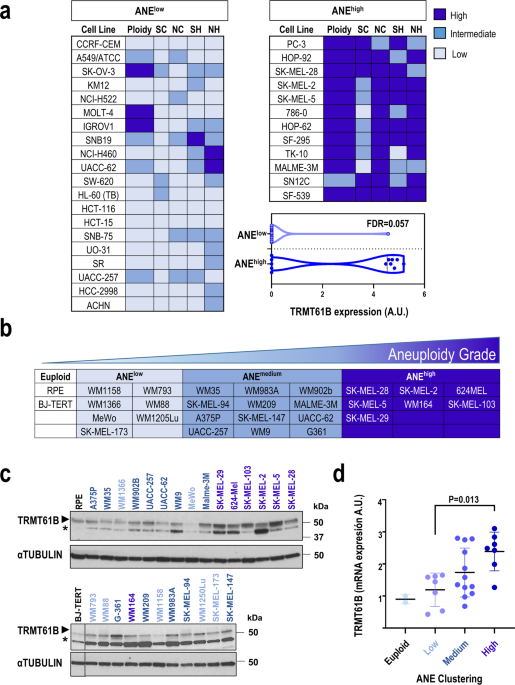
To discover novel molecular traits related to high aneuploidy, we used publicly available data on the NCI-60 human cancer cell line panel [39,40,41,42,43,44,45,46]. Using DNA ploidy
information and 4 different structural and numerical karyotypic features (Supplementary Table 1) [44], we performed unsupervised hierarchical analysis to stratify the NCI-60 cell lines
included in the panel into 3 groups: low, intermediate, and high aneuploidy levels (Sup. Figure 1a). We compared the proteomic signatures [43] of the group with the highest (ANEhigh) vs. the
lowest (ANElow) aneuploidy levels (Fig. 1a) to identify proteins strongly associated with aneuploidy. Taking FDR