Early immune modulation by single-agent trastuzumab as a marker of trastuzumab benefit
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Optimising the selection of HER2-targeted regimens by identifying subsets of HER2-positive breast cancer (BC) patients who need more or less therapy remains challenging. We analysed BC
samples before and after treatment with 1 cycle of trastuzumab according to the response to trastuzumab.
Gene expression profiles of pre- and post-treatment tumour samples from 17 HER2-positive BC patients were analysed on the Illumina platform. Tumour-associated immune pathways and blood
counts were analysed with regard to the response to trastuzumab. HER2-positive murine models with differential responses to trastuzumab were used to reproduce and better characterise these
data.
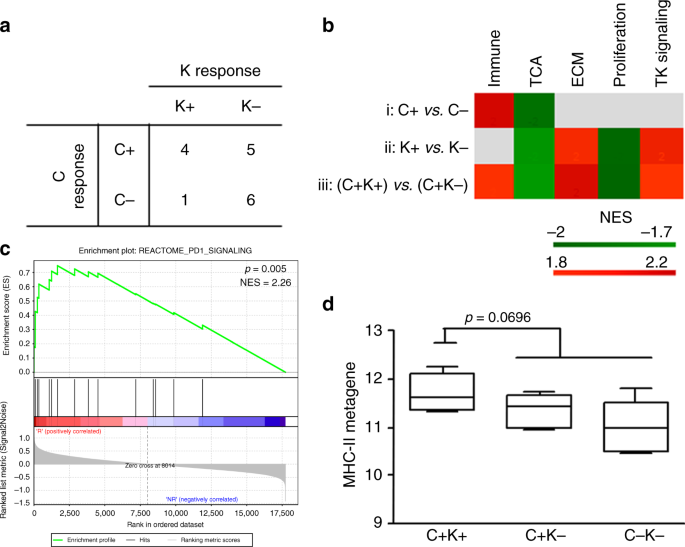
Patients who responded to single-agent trastuzumab had basal tumour biopsies that were enriched in immune pathways, particularly the MHC-II metagene. One cycle of trastuzumab modulated the
expression levels of MHC-II genes, which increased in patients who had a complete response on treatment with trastuzumab and chemotherapy. Trastuzumab increased the MHC-II-positive cell
population, primarily macrophages, only in the tumour microenvironment of responsive mice. In patients who benefited from complete trastuzumab therapy and in mice that harboured responsive
tumours circulating neutrophil levels declined, but this cell subset rose in nonresponsive tumours.
Short treatment with trastuzumab induces local and systemic immunomodulation that is associated with clinical outcomes.
The HER2 receptor is overexpressed in approximately 20% of breast cancers (BCs) and has historically been associated with aggressive disease and a poor prognosis. Treatment with trastuzumab
and chemotherapy has dramatically changed the prognosis of early and advanced HER2-positive (HER2+) BC patients.1 More recently, the introduction and availability of other HER2-targeting
agents offer clinicians the ability to provide prolonged inhibition of HER2 signalling across multiple lines of treatment, and the dual HER2 blockade approach has demonstrated superior
activity over trastuzumab alone. As more potential therapies appear over the horizon, advancements in biomarker discovery will be critical in optimising treatment selection and providing
personalised therapy for patients. Particularly, it remains unknown which patients benefit from single-agent trastuzumab and those who require instead the dual blockade upfront. Many efforts
have been made toward identifying biomarkers that predict a benefit from trastuzumab: HER2 addiction and immune features have demonstrated the best predictive ability in several trials
(reviewed in ref. 2), but HER2 expression in the primary tumour remains the only marker that is used in clinical practice.
Preclinical studies have reported that trastuzumab has cytostatic (e.g., inhibition of HER2 signalling, extracellular domain shedding, and tumour angiogenesis) and cytotoxic activities
[antibody-dependent cell-mediated cytotoxicity (ADCC), antibody-dependent cell-mediated phagocytosis (ADCP), and inhibition of DNA repair].3 In the neoadjuvant setting, the decrease in
tumour burden, or pathological complete response (pCR), is likely to depend on the cytotoxic activity of trastuzumab.4 Accordingly, increasing evidence has shown that the therapeutic
activity of trastuzumab relies on the innate and adaptive immune systems, and the significant pre-existing infiltration of BCs by immune cells has been associated with pCR in several trials
(reviewed in ref. 5). Trastuzumab also positively regulates infiltration by lymphoid cells in tumour tissues,6,7,8 supporting that its clinical activity is based on its immunostimulatory
effects.9
In this study, we analysed the gene expression profile of tumour biopsies from the TRastuzumab UPfront in HER2+ locally advanced BC (TRUP) window-of-opportunity trial10 before and after
brief exposure to trastuzumab. We performed an exploratory analysis of the molecular features that were associated with the clinical activity of short-term trastuzumab monotherapy,
monitoring changes in immune expression on treatment as markers of trastuzumab activity.
The 17 tumours that we profiled were obtained from BC patients in the TRUP window-of-opportunity trial,10 which comprised 28 locally advanced primary BCs that were diagnosed by incisional
biopsy at A.O. Istituti Ospitalieri di Cremona. In the TRUP study, patients received 1 cycle of trastuzumab alone, followed by 4 cycles of trastuzumab and paclitaxel, until definitive
surgery. As per the protocol, after single-agent trastuzumab (21 days), assessment of tumour response and research tumour biopsy (Tru-cut biopsy) were performed. Responses at the Tru-cut
biopsy were available for all but one patient. Blood test results (i.e., blood cell count/μl) at baseline and at the Tru-cut biopsy were retrieved and analysed. All procedures were performed
in accordance with the Helsinki Declaration. The biospecimens that were examined consisted of leftover material from samples that had been collected during standard surgical and medical
procedures at A.O. Istituti Ospitalieri di Cremona. Samples were donated by patients to the Institutional BioBank for research purposes, and aliquots were designated for this study after
approval by the institutional review board and a specific request to the independent ethical committee ‘Comitato Etico Val Padana’.
The response to trastuzumab after 21 days was calculated as % reduction in clinical dimensions or in the number of Ki67-positive cells at day 21 compared with baseline. Patients who
experienced a decrease of at least 20% in tumour volume or at least 50% in the number of Ki67-positive cells were scored as responders. The response rate (RR) to trastuzumab and chemotherapy
was calculated as the ratio of the difference between the initial clinical dimensions and final pathological dimensions to the initial clinical dimensions, and responders were selected,
based on the cut-off RR in the original study (RR = 70%).10
RNA was extracted from frozen incisional biopsies of the TRUP cohort using the miRNeasy MINI KIT (Qiagen) according to the manufacturer’s protocol. RNA quality was checked in a preanalytical
screen by RT-qPCR, as described.11 Gene expression profiles were generated by whole-genome DASL (cDNA-mediated annealing, selection, extension, and ligation) assay using HumanHT12_v4
BeadChips (Illumina), according to a standard protocol. The Illumina BeadArray Reader was used to scan the arrays. Illumina BeadScan was used to acquire the images and recover the primary
data. The data were quantile-normalised using BeadStudio. The BeadChips covered over 29,000 annotated genes that were derived from RefSeq (Build 36.2, Release 38), and after a filter step, a
data matrix that contained 17 matched samples was generated. The data were deposited into the Gene Expression Omnibus repository (accession number GSE114082).
Gene set enrichment analysis was performed for Reactome pathways using GSEA v2.0.13.12 Genes that were represented by more than 1 probe were collapsed to the probe with the maximum value
using the Collapse Dataset tool. Gene set permutation type was applied 1000 times, and gene set enrichment was considered to be significant at a false discovery rate (FDR) of