The pol ii ctd: new twists in the tail
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
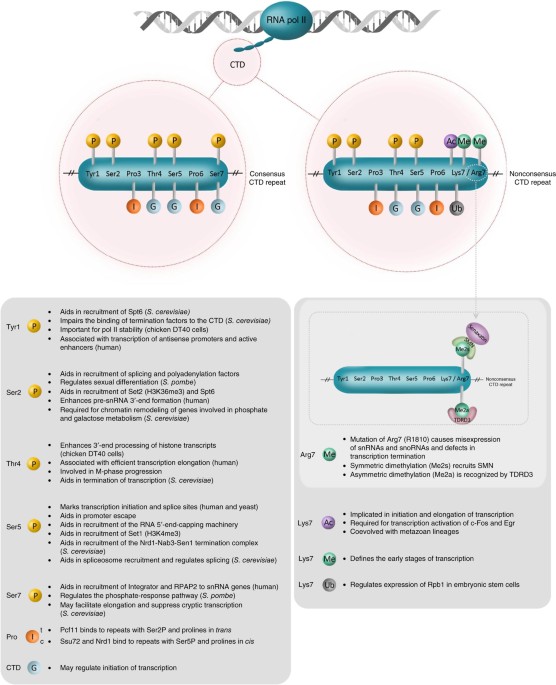
ABSTRACT The C-terminal domain (CTD) of the large subunit of RNA polymerase (pol) II comprises conserved heptad repeats, and post-translational modification of the CTD regulates
transcription and cotranscriptional RNA processing. Recently, the spatial patterns of modification of the CTD repeats have been investigated, and new functions of CTD modification have been
revealed. In addition, there are new insights into the roles of the enzymes that decorate the CTD. We review these new findings and reassess the role of the pol II CTD in the regulation of
gene expression. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution
Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full
article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs *
Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS STRUCTURAL INSIGHTS INTO NUCLEAR TRANSCRIPTION BY EUKARYOTIC DNA-DEPENDENT RNA POLYMERASES Article 03 May 2022 STRUCTURAL
INSIGHTS INTO TRANSCRIPTIONAL REGULATION OF HUMAN RNA POLYMERASE III Article 08 February 2021 STRUCTURAL INSIGHTS INTO HUMAN POL III TRANSCRIPTION INITIATION IN ACTION Article 04 June 2025
REFERENCES * Eick, D. & Geyer, M. The RNA polymerase II carboxy-terminal domain (CTD) code. _Chem. Rev._ 113, 8456–8490 (2013). CAS PubMed Google Scholar * Jeronimo, C., Collin, P.
& Robert, F. The RNA polymerase II CTD: the increasing complexity of a low-complexity protein domain. _J. Mol. Biol._ 428, 2607–2622 (2016). CAS PubMed Google Scholar * Srivastava, R.
& Ahn, S.H. Modifications of RNA polymerase II CTD: connections to the histone code and cellular function. _Biotechnol. Adv._ 33, 856–872 (2015). CAS PubMed Google Scholar * Egloff,
S., Dienstbier, M. & Murphy, S. Updating the RNA polymerase CTD code: adding gene-specific layers. _Trends Genet._ 28, 333–341 (2012). Article CAS PubMed Google Scholar * Voss, K. et
al. Site-specific methylation and acetylation of lysine residues in the C-terminal domain (CTD) of RNA polymerase II. _Transcription_ 6, 91–101 (2015). CAS PubMed PubMed Central Google
Scholar * Dias, J.D. et al. Methylation of RNA polymerase II non-consensus lysine residues marks early transcription in mammalian cells. _eLife_ 4, e11215 (2015). PubMed PubMed Central
Google Scholar * Li, H. et al. Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. _Mol. Cell. Biol._ 27, 5296–5305 (2007). CAS
PubMed PubMed Central Google Scholar * Sims, R.J. III. et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. _Science_ 332, 99–103 (2011). CAS
PubMed PubMed Central Google Scholar * Zhao, D.Y. et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. _Nature_ 529, 48–53 (2016).
PubMed Google Scholar * Hintermair, C. et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. _EMBO J._ 31,
2784–2797 (2012). CAS PubMed PubMed Central Google Scholar * Descostes, N. et al. Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription
and active enhancers in mammalian cells. _eLife_ 3, e02105 (2014). PubMed PubMed Central Google Scholar * Nojima, T. et al. Mammalian NET-seq reveals genome-wide nascent transcription
coupled to RNA processing. _Cell_ 161, 526–540 (2015). CAS PubMed PubMed Central Google Scholar * Milligan, L. et al. Strand-specific, high-resolution mapping of modified RNA polymerase
II. _Mol. Syst. Biol._ 12, 874 (2016). PubMed PubMed Central Google Scholar * Churchman, L.S. & Weissman, J.S. Nascent transcript sequencing visualizes transcription at nucleotide
resolution. _Nature_ 469, 368–373 (2011). CAS PubMed Google Scholar * Lu, L. et al. Distributive O-GlcNAcylation on the highly repetitive C-terminal domain of RNA polymerase II.
_Biochemistry_ 55, 1149–1158 (2016). CAS PubMed Google Scholar * Mayer, A. et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. _Science_ 336,
1723–1725 (2012). CAS PubMed Google Scholar * Harlen, K.M. et al. Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho-CTD residue. _Cell Rep._
15, 2147–2158 (2016). CAS PubMed PubMed Central Google Scholar * Bataille, A.R. et al. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase,
phosphatase, and isomerase enzymes along genes. _Mol. Cell_ 45, 158–170 (2012). CAS PubMed Google Scholar * Mayer, A. et al. Uniform transitions of the general RNA polymerase II
transcription complex. _Nat. Struct. Mol. Biol._ 17, 1272–1278 (2010). CAS PubMed Google Scholar * Tietjen, J.R. et al. Chemical-genomic dissection of the CTD code. _Nat. Struct. Mol.
Biol._ 17, 1154–1161 (2010). CAS PubMed PubMed Central Google Scholar * Kim, H. et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. _Nat. Struct. Mol. Biol._ 17,
1279–1286 (2010). CAS PubMed PubMed Central Google Scholar * Suh, H. et al. Direct analysis of phosphorylation sites on the Rpb1 C-terminal domain of RNA polymerase II. _Mol. Cell_ 61,
297–304 (2016). CAS PubMed PubMed Central Google Scholar * Schüller, R. et al. Heptad-specific phosphorylation of RNA P=polymerase II CTD. _Mol. Cell_ 61, 305–314 (2016). PubMed Google
Scholar * Stiller, J.W. & Cook, M.S. Functional unit of the RNA polymerase II C-terminal domain lies within heptapeptide pairs. _Eukaryot. Cell_ 3, 735–740 (2004). CAS PubMed PubMed
Central Google Scholar * Akhtar, M.S. et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. _Mol. Cell_ 34, 387–393 (2009). CAS PubMed PubMed
Central Google Scholar * Glover-Cutter, K. et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination
by RNA polymerase II. _Mol. Cell. Biol._ 29, 5455–5464 (2009). CAS PubMed PubMed Central Google Scholar * Galbraith, M.D., Donner, A.J. & Espinosa, J.M. CDK8: a positive regulator of
transcription. _Transcription_ 1, 4–12 (2010). CAS PubMed PubMed Central Google Scholar * Galbraith, M.D. et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response
to hypoxia. _Cell_ 153, 1327–1339 (2013). CAS PubMed PubMed Central Google Scholar * Poss, Z.C. et al. Identification of Mediator kinase substrates in human cells using cortistatin a and
quantitative phosphoproteomics. _Cell Rep._ 15, 436–450 (2016). CAS PubMed PubMed Central Google Scholar * Bartkowiak, B. et al. CDK12 is a transcription elongation-associated CTD
kinase, the metazoan ortholog of yeast Ctk1. _Genes Dev._ 24, 2303–2316 (2010). Article CAS PubMed PubMed Central Google Scholar * Pei, Y., Schwer, B. & Shuman, S. Interactions
between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. _J. Biol. Chem._ 278, 7180–7188 (2003). CAS
PubMed Google Scholar * Karagiannis, J. & Balasubramanian, M.K. A cyclin-dependent kinase that promotes cytokinesis through modulating phosphorylation of the carboxy terminal domain of
the RNA Pol II Rpb1p sub-unit. _PLoS One_ 2, e433 (2007). PubMed PubMed Central Google Scholar * Laitem, C. et al. CDK9 inhibitors define elongation checkpoints at both ends of RNA
polymerase II–transcribed genes. _Nat. Struct. Mol. Biol._ 22, 396–403 (2015). CAS PubMed PubMed Central Google Scholar * Greifenberg, A.K. et al. Structural and functional analysis of
the Cdk13/Cyclin K complex. _Cell Rep._ 14, 320–331 (2016). CAS PubMed Google Scholar * Ghamari, A. et al. _In vivo_ live imaging of RNA polymerase II transcription factories in primary
cells. _Genes Dev._ 27, 767–777 (2013). CAS PubMed PubMed Central Google Scholar * Czudnochowski, N., Bösken, C.A. & Geyer, M. Serine-7 but not serine-5 phosphorylation primes RNA
polymerase II CTD for P-TEFb recognition. _Nat. Commun._ 3, 842 (2012). PubMed Google Scholar * Blazek, D. et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of
expression of DNA damage response genes. _Genes Dev._ 25, 2158–2172 (2011). CAS PubMed PubMed Central Google Scholar * Bowman, E.A., Bowman, C.R., Ahn, J.H. & Kelly, W.G.
Phosphorylation of RNA polymerase II is independent of P-TEFb in the _C. elegans_ germline. _Development_ 140, 3703–3713 (2013). CAS PubMed PubMed Central Google Scholar * Cheng, S.W. et
al. Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. _Mol. Cell. Biol._ 32, 4691–4704
(2012). CAS PubMed PubMed Central Google Scholar * Davidson, L., Muniz, L. & West, S. 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are
reciprocally coupled in human cells. _Genes Dev._ 28, 342–356 (2014). CAS PubMed PubMed Central Google Scholar * Eifler, T.T. et al. Cyclin-dependent kinase 12 increases 3′ end
processing of growth factor-induced c-FOS transcripts. _Mol. Cell. Biol._ 35, 468–478 (2015). PubMed Google Scholar * Yu, M. et al. RNA polymerase II-associated factor 1 regulates the
release and phosphorylation of paused RNA polymerase II. _Science_ 350, 1383–1386 (2015). CAS PubMed PubMed Central Google Scholar * Liang, K. et al. Characterization of human
cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. _Mol. Cell. Biol._ 35, 928–938 (2015). CAS PubMed
PubMed Central Google Scholar * Bartkowiak, B., Yan, C. & Greenleaf, A.L. Engineering an analog-sensitive CDK12 cell line using CRISPR/Cas. _Biochim. Biophys. Acta_ 1849, 1179–1187
(2015). CAS PubMed PubMed Central Google Scholar * Bösken, C.A. et al. The structure and substrate specificity of human Cdk12/Cyclin K. _Nat. Commun._ 5, 3505 (2014). PubMed Google
Scholar * Di Vona, C. et al. Chromatin-wide profiling of DYRK1A reveals a role as a gene-specific RNA polymerase II CTD kinase. _Mol. Cell_ 57, 506–520 (2015). CAS PubMed Google Scholar
* Pak, V. et al. CDK11 in TREX/THOC regulates HIV mRNA 3′ end processing. _Cell Host Microbe_ 18, 560–570 (2015). CAS PubMed PubMed Central Google Scholar * Devaiah, B.N. et al. BRD4 is
an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. _Proc. Natl. Acad. Sci. USA_ 109, 6927–6932 (2012). CAS PubMed PubMed Central Google
Scholar * Hsin, J.P., Sheth, A. & Manley, J.L. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. _Science_ 334, 683–686 (2011). CAS PubMed
PubMed Central Google Scholar * Baskaran, R., Escobar, S.R. & Wang, J.Y. Nuclear c-Abl is a COOH-terminal repeated domain (CTD)-tyrosine (CTD)-tyrosine kinase-specific for the
mammalian RNA polymerase II: possible role in transcription elongation. _Cell Growth Differ._ 10, 387–396 (1999). CAS PubMed Google Scholar * Schröder, S. et al. Acetylation of RNA
polymerase II regulates growth-factor-induced gene transcription in mammalian cells. _Mol. Cell_ 52, 314–324 (2013). PubMed PubMed Central Google Scholar * Mosley, A.L. et al. Rtr1 is a
CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. _Mol. Cell_ 34, 168–178 (2009). CAS PubMed PubMed Central Google Scholar
* Egloff, S., Zaborowska, J., Laitem, C., Kiss, T. & Murphy, S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. _Mol. Cell_ 45, 111–122 (2012). CAS
PubMed PubMed Central Google Scholar * Ni, Z. et al. RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. _Nat. Struct. Mol. Biol._ 21,
686–695 (2014). CAS PubMed PubMed Central Google Scholar * Krishnamurthy, S., He, X., Reyes-Reyes, M., Moore, C. & Hampsey, M. Ssu72 Is an RNA polymerase II CTD phosphatase. _Mol.
Cell_ 14, 387–394 (2004). CAS PubMed Google Scholar * Zhang, D.W. et al. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential
for viability and transcription termination. _J. Biol. Chem._ 287, 8541–8551 (2012). CAS PubMed PubMed Central Google Scholar * O'Reilly, D. et al. Human snRNA genes use
polyadenylation factors to promote efficient transcription termination. _Nucleic Acids Res._ 42, 264–275 (2014). CAS PubMed Google Scholar * Cho, E.J., Kobor, M.S., Kim, M., Greenblatt,
J. & Buratowski, S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. _Genes Dev._ 15, 3319–3329 (2001). CAS PubMed PubMed
Central Google Scholar * Schreieck, A. et al. RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. _Nat. Struct. Mol. Biol._ 21, 175–179
(2014). CAS PubMed PubMed Central Google Scholar * Hsu, P.L. et al. Rtr1 is a dual specificity phosphatase that dephosphorylates Tyr1 and Ser5 on the RNA polymerase II CTD. _J. Mol.
Biol._ 426, 2970–2981 (2014). CAS PubMed PubMed Central Google Scholar * McCracken, S. et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription.
_Nature_ 385, 357–361 (1997). CAS PubMed Google Scholar * Grzechnik, P., Gdula, M.R. & Proudfoot, N.J. Pcf11 orchestrates transcription termination pathways in yeast. _Genes Dev._ 29,
849–861 (2015). CAS PubMed PubMed Central Google Scholar * Burugula, B.B. et al. Histone deacetylases and phosphorylated polymerase II C-terminal domain recruit Spt6 for
cotranscriptional histone reassembly. _Mol. Cell. Biol._ 34, 4115–4129 (2014). PubMed PubMed Central Google Scholar * Hintermair, C. et al. Specific threonine-4 phosphorylation and
function of RNA polymerase II CTD during M phase progression. _Sci. Rep._ 6, 27401 (2016). CAS PubMed PubMed Central Google Scholar * Simonti, C.N. et al. Evolution of lysine acetylation
in the RNA polymerase II C-terminal domain. _BMC Evol. Biol._ 15, 35 (2015). PubMed PubMed Central Google Scholar * Ranuncolo, S.M., Ghosh, S., Hanover, J.A., Hart, G.W. & Lewis,
B.A. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription _in vitro_ and _in vivo_. _J. Biol. Chem._ 287, 23549–23561 (2012). CAS PubMed PubMed
Central Google Scholar * Brès, V., Yoh, S.M. & Jones, K.A. The multi-tasking P-TEFb complex. _Curr. Opin. Cell Biol._ 20, 334–340 (2008). PubMed PubMed Central Google Scholar *
Zaborowska, J., Isa, N.F. & Murphy, S. P-TEFb goes viral. _Inside Cell_ 1, 106–116 (2016). PubMed Google Scholar * Kwak, H. & Lis, J.T. Control of transcriptional elongation.
_Annu. Rev. Genet._ 47, 483–508 (2013). CAS PubMed PubMed Central Google Scholar * O'Brien, S.K., Cao, H., Nathans, R., Ali, A. & Rana, T.M. P-TEFb kinase complex phosphorylates
histone H1 to regulate expression of cellular and HIV-1 genes. _J. Biol. Chem._ 285, 29713–29720 (2010). CAS PubMed PubMed Central Google Scholar * Kim, J.B. & Sharp, P.A. Positive
transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. _J. Biol. Chem._ 276,
12317–12323 (2001). CAS PubMed Google Scholar * Shchebet, A., Karpiuk, O., Kremmer, E., Eick, D. & Johnsen, S.A. Phosphorylation by cyclin-dependent kinase-9 controls
ubiquitin-conjugating enzyme-2A function. _Cell Cycle_ 11, 2122–2127 (2012). CAS PubMed Google Scholar * Sansó, M. et al. P-TEFb regulation of transcription termination factor Xrn2
revealed by a chemical genetic screen for Cdk9 substrates. _Genes Dev._ 30, 117–131 (2016). PubMed PubMed Central Google Scholar * Chen, F.X. et al. PAF1, a molecular regulator of
promoter-proximal pausing by RNA polymerase II. _Cell_ 162, 1003–1015 (2015). CAS PubMed PubMed Central Google Scholar * Chathoth, K.T., Barrass, J.D., Webb, S. & Beggs, J.D. A
splicing-dependent transcriptional checkpoint associated with prespliceosome formation. _Mol. Cell_ 53, 779–790 (2014). CAS PubMed PubMed Central Google Scholar * Fong, Y.W. & Zhou,
Q. Stimulatory effect of splicing factors on transcriptional elongation. _Nature_ 414, 929–933 (2001). CAS PubMed Google Scholar * Koga, M., Hayashi, M. & Kaida, D. Splicing
inhibition decreases phosphorylation level of Ser2 in Pol II CTD. _Nucleic Acids Res._ 43, 8258–8267 (2015). CAS PubMed PubMed Central Google Scholar * Yoh, S.M., Cho, H., Pickle, L.,
Evans, R.M. & Jones, K.A. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. _Genes Dev._ 21, 160–174 (2007). CAS PubMed PubMed Central Google
Scholar * Vasiljeva, L., Kim, M., Mutschler, H., Buratowski, S. & Meinhart, A. The Nrd1–Nab3–Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal
domain. _Nat. Struct. Mol. Biol._ 15, 795–804 (2008). CAS PubMed PubMed Central Google Scholar * Noble, C.G. et al. Key features of the interaction between Pcf11 CID and RNA polymerase
II CTD. _Nat. Struct. Mol. Biol._ 12, 144–151 (2005). CAS PubMed Google Scholar * Werner-Allen, J.W. et al. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II
C-terminal domain phosphatase Ssu72. _J. Biol. Chem._ 286, 5717–5726 (2011). CAS PubMed Google Scholar * Kubicek, K. et al. Serine phosphorylation and proline isomerization in RNAP II CTD
control recruitment of Nrd1. _Genes Dev._ 26, 1891–1896 (2012). CAS PubMed PubMed Central Google Scholar * Rosonina, E. et al. Threonine-4 of the budding yeast RNAP II CTD couples
transcription with Htz1-mediated chromatin remodeling. _Proc. Natl. Acad. Sci. USA_ 111, 11924–11931 (2014). CAS PubMed PubMed Central Google Scholar * Lewis, B.A. O-GlcNAcylation at
promoters, nutrient sensors, and transcriptional regulation. _Biochim. Biophys. Acta_ 1829, 1202–1206 (2013). CAS PubMed Google Scholar * Chatterjee, D., Sanchez, A.M., Goldgur, Y.,
Shuman, S. & Schwer, B. Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho-sites govern the repression of pho1 gene expression under
phosphate-replete conditions in fission yeast. _RNA_ 22, 1011–1025 (2016). CAS PubMed PubMed Central Google Scholar * Coudreuse, D. et al. A gene-specific requirement of RNA polymerase
II CTD phosphorylation for sexual differentiation in _S. pombe_. _Curr. Biol._ 20, 1053–1064 (2010). CAS PubMed Google Scholar * Clemente-Blanco, A. et al. Cdc14 phosphatase promotes
segregation of telomeres through repression of RNA polymerase II transcription. _Nat. Cell Biol._ 13, 1450–1456 (2011). CAS PubMed PubMed Central Google Scholar * Kim, M., Suh, H., Cho,
E.J. & Buratowski, S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. _J. Biol. Chem._ 284, 26421–26426 (2009). CAS PubMed PubMed Central Google Scholar *
Zhang, M. et al. Structural and kinetic analysis of prolyl-isomerization/phosphorylation cross-talk in the CTD code. _ACS Chem. Biol._ 7, 1462–1470 (2012). CAS PubMed PubMed Central
Google Scholar * Wani, S., Sugita, A., Ohkuma, Y. & Hirose, Y. Human SCP4 is a chromatin-associated CTD phosphatase and exhibits the dynamic translocation during erythroid
differentiation. _J. Biochem._ 160, 111–120 (2016). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank L. Vasiljeva, N. Proudfoot, M. Tellier and I. Ferrer-Vicens for
discussions and critical comments. The authors also thank F. Vazquez-Arango and M. Tellier for assistance with generating figures. This work was supported by grants from the Wellcome Trust
(WT106134A) to S.M. and by Fondation ARC to S.E. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Sir William Dunn School of Pathology, University of Oxford, Oxford, UK Justyna Zaborowska &
Shona Murphy * Laboratoire de Biologie Moléculaire Eucaryote, CNRS, Université Paul Sabatier, Toulouse, France Sylvain Egloff Authors * Justyna Zaborowska View author publications You can
also search for this author inPubMed Google Scholar * Sylvain Egloff View author publications You can also search for this author inPubMed Google Scholar * Shona Murphy View author
publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Shona Murphy. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing financial interests. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Zaborowska, J., Egloff, S. & Murphy, S. The pol II CTD: new twists in
the tail. _Nat Struct Mol Biol_ 23, 771–777 (2016). https://doi.org/10.1038/nsmb.3285 Download citation * Received: 07 March 2016 * Accepted: 03 August 2016 * Published: 06 September 2016 *
Issue Date: September 2016 * DOI: https://doi.org/10.1038/nsmb.3285 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative