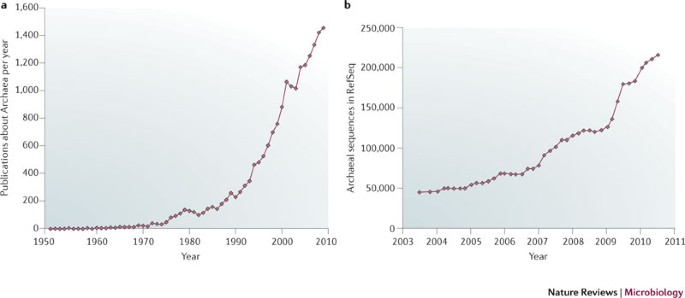
Archaea — timeline of the third domain
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
KEY POINTS * Studies of the Archaea have had a substantial impact on the field of biology. * Carl Woese carried out several pioneering studies, examining the evolution of the genetic code,
the translation apparatus and the cell as a whole, and the insight gained from his early work led to the discovery of the third domain. * Defining moments in the history of archaeal research
include the construction of a universal tree of life and the rationalization of the phylogeny of small-subunit ribosomal RNA, aminoacyl-tRNA synthetases and other protein sequences, leading
to the three-domain view of life. * Seminal advances were made through probing the biochemistry, physiology, genetics and evolution of methanogens, extreme halophiles and thermoacidophiles.
* The nature of archaea as extremophiles can now be placed into context with the knowledge that archaea are abundant and ubiquitous throughout the Earth's biosphere, including in the
vast cold reaches of the planet. * Archaea play a key part in maintaining important biogeochemical cycles, and several contemporary advances have been made in our understanding of the
importance of archaea in global ecology. * Technological advances have greatly enhanced the output from the field; particularly noteworthy are the impact of DNA sequencing, and the dawning
of the genomics era and application of metagenomics, as well as breakthroughs that have been made through the development of tractable genetic systems. ABSTRACT The Archaea evolved as one of
the three primary lineages several billion years ago, but the first archaea to be discovered were described in the scientific literature about 130 years ago. Moreover, the Archaea were
formally proposed as the third domain of life only 20 years ago. Over this very short period of investigative history, the scientific community has learned many remarkable things about the
Archaea — their unique cellular components and pathways, their abundance and critical function in diverse natural environments, and their quintessential role in shaping the evolutionary path
of life on Earth. This Review charts the 'archaea movement', from its genesis through to key findings that, when viewed together, illustrate just how strongly the field has built
on new knowledge to advance our understanding not only of the Archaea, but of biology as a whole. Access through your institution Buy or subscribe This is a preview of subscription content,
access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn
more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS
OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS THE CELL BIOLOGY OF ARCHAEA Article 17 October
2022 THE EMERGING VIEW ON THE ORIGIN AND EARLY EVOLUTION OF EUKARYOTIC CELLS Article 11 September 2024 CO‐EVOLUTION OF EARLY EARTH ENVIRONMENTS AND MICROBIAL LIFE Article 29 May 2024
REFERENCES * Cavicchioli, R. (ed) _Archaea: Molecular and Cellular Biology_ (American Society for Microbiology Press, Washington DC, 2007). A STRONG COLLECTION OF CHAPTERS FROM LEADERS IN
THE FIELD, INCLUDING A TESTAMENT ABOUT THE DISCOVERY OF THE ARCHAEA BY CARL WOESE. Book Google Scholar * Garrett, R. & Klenk, H.-P. _Archaea: Evolution, Physiology and Molecular
Biology_ (Blackwell, Oxford, UK, 2007). Google Scholar * Rothschild, L. J. & Mancinelli, R. L. Life in extreme environments. _Nature_ 409, 1092–1101 (2001). Article CAS PubMed Google
Scholar * Kletzin, A. in _Archaea: Molecular and Cellular Biology_ (ed. Cavicchioli, R.) 14–92 (American Society for Microbiology Press, Washington DC, 2007). A COMPREHENSIVE OVERVIEW OF
THE MICROBIOLOGY, BIOCHEMISTRY AND ECOLOGY OF MANY TYPES OF ARCHAEA. Book Google Scholar * Makarova, K. S., Yutin, N., Bell, S. D. & Koonin, E. V. Evolution of diverse cell division
and vesicle formation systems in Archaea. _Nature Rev. Microbiol._ 8, 731–741 (2010). Article CAS Google Scholar * Gribaldo, S., Poole, A. M., Daubin, V., Forterre, P. &
Brochier-Armanet, C. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? _Nature Rev. Microbiol._ 8, 743–752 (2010). Article CAS Google
Scholar * Walsby, A. E. A square bacterium. _Nature_ 283, 69–71 (1980). Article Google Scholar * Burns, D. G. et al. _Haloquadratum walsbyi_ gen. nov., sp. nov., the square haloarchaeon
of Walsby, isolated from saltern crystallizers in Australia and Spain. _Int. J. Syst. Evol. Microbiol._ 57, 387–392 (2007). Article CAS PubMed Google Scholar * Soppa, J. From genomes to
function: haloarchaea as model organisms. _Microbiology_ 152, 585–590 (2006). Article CAS PubMed Google Scholar * Cavicchioli, R., Curmi, P. M. G., Saunders, N. & Thomas, T.
Pathogenic archaea: do they exist? _Bioessays_ 25, 1119–1128 (2003). A DISCUSSION ABOUT THE CONCEPT OF ARCHAEAL PATHOGENS. Article CAS PubMed Google Scholar * Lepp, P. W. et al.
Methanogenic Archaea and human periodontal disease. _Proc. Natl Acad. Sci. USA_ 101, 6176–6181 (2004). Article CAS PubMed PubMed Central Google Scholar * Qin, J. et al. A human gut
microbial gene catalogue established by metagenomic sequencing. _Nature_ 464, 59–65 (2010). Article CAS PubMed PubMed Central Google Scholar * Brulc, J. M. et al. Gene-centric
metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. _Proc. Natl Acad. Sci. USA_ 106, 1948–1953 (2009). Article PubMed PubMed Central
Google Scholar * Burkhardt, F. & Smith, S. (eds) _The Correspondence of Charles Darwin:1856–1857_ Vol. 6 (Cambridge Univ. Press, Cambridge, UK, 1990). Google Scholar * Woese, C. R. in
_Organization and Control in Prokaryotic and Eukaryotic Cells. 20th Symposium, Society for General Microbiology_ (eds Charles, H. P. & Knight, B. C. J. G.) 39–54 (Cambridge Univ. Press,
Cambridge, UK (1970). Google Scholar * Woese, C. R., Sogin, M. L. & Sutton, L. A. Procaryote phylogeny. I. Concerning the relatedness of _Aerobacter aerogenes_ to _Escherichia coli_.
_J. Mol. Evol._ 3, 293–299 (1974). Article CAS PubMed Google Scholar * Crick, F. H. C. The biological replication of macromolecules. _Symp. Soc. Exp. Biol._ 12, 138–163 (1958). CAS
PubMed Google Scholar * Zuckerkandl, E. & Pauling, L. Molecules as documents of evolutionary history. _J. Theor. Biol._ 8, 357–366 (1965). Article CAS PubMed Google Scholar * Fox,
G. E., Magrum, L. J., Balch, W. E., Wolfe, R. S. & Woese, C. R. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. _Proc. Natl Acad. Sci. USA_ 74, 4537–4541
(1977). A CORNERSTONE OF THE DISCOVERY OF THE THIRD DOMAIN: THE FIRST SSU RRNA SEQUENCES OF METHANOGENS. Article CAS PubMed PubMed Central Google Scholar * Woese, C. R. & Fox, G. E.
The phylogenetic structure of the procaryotic domain: the primary kingdoms. _Proc. Natl Acad. Sci. USA_ 74, 5088–5090 (1977). THE QUINTESSENTIAL DESCRIPTION OF THE THIRD DOMAIN THAT STARTED
IT ALL. Article CAS PubMed PubMed Central Google Scholar * Woese, C. R. & Fox, G. E. The concept of cellular evolution. _J. Mol. Evol._ 10, 1–6 (1977). An important paper
describing the importance of the genotype–phenotype relationship in the evolution of life. Article CAS PubMed Google Scholar * Woese, C. R., Kandler, O. & Wheelis, M. L. Towards a
natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. _Proc. Natl Acad. Sci. USA_ 87, 4576–4579 (1990). THE PROPOSAL FOR THE FORMAL DESCRIPTION OF THE THREE
DOMAINS — A MUST-READ FOR ALL EDUCATIONAL PURPOSES. Article CAS PubMed PubMed Central Google Scholar * Woese, C. R. in _Archaea: Molecular and Cellular Biology_ (ed. Cavicchioli, R.)
1–13 (American Society for Microbiology Press, Washington DC, 2007). A PROVOCATIVE TESTAMENT ABOUT THE ARCHAEA MOVEMENT. Book Google Scholar * Cavalier-Smith, T. The neomuran origin of
archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. _Int. J. Syst. Evol. Microbiol._ 52, 7–76 (2002). Article CAS PubMed Google Scholar *
Simonson, A. B. et al. Decoding the genomic tree of life. _Proc. Natl Acad. Sci. USA_ 102, 6608–6613 (2005). Article CAS PubMed PubMed Central Google Scholar * Sapp, J. The
prokaryote-eukaryote dichotomy: meanings and mythology. _Microbiol. Mol. Biol. Rev._ 69, 292–305 (2005). AN EXTENSIVE AND REVEALING REVIEW ABOUT THE ARCHAEA MOVEMENT. Article CAS PubMed
PubMed Central Google Scholar * Friend, T. _The Third Domain: the Untold Story of Archaea and the Future of Biotechnology_. (Joseph Henry, Washington DC, 2007). AN ENJOYABLE, EASY-TO-READ
AND INSIGHTFUL BOOK ABOUT THE HISTORY OF THE DISCOVERY OF THE ARCHAEA. Google Scholar * Woese, C. R. & Goldenfeld, N. How the microbial world saved evolution from the Scylla of
molecular biology and the Charybdis of the modern synthesis. _Microbiol. Mol. Biol. Rev._ 73, 14–21 (2009). A CONTEMPORARY ESSAY CONCERNING HOW THE EVOLUTIONARY PROCESS HAS BEEN STUDIED IN
THE TWENTIETH CENTURY. Article PubMed PubMed Central Google Scholar * Iwabe, N., Kuma, K., Hasegawa, M., Osawa, S. & Miyata, T. Evolutionary relationship of archaebacteria,
eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. _Proc. Natl Acad. Sci. USA_ 86, 9355–9359 (1989). Article CAS PubMed PubMed Central Google Scholar *
Brown, J. R. & Doolittle, W. F. Archaea and the prokaryote-to-eukaryote transition. _Microbiol. Mol. Biol. Rev._ 61, 456–502 (1997). AN EXTENSIVE REVIEW OF THE EVOLUTION OF THE UNIVERSAL
TREE OF LIFE, WITH PARTICULAR EMPHASIS ON PROTEIN PHYLOGENY. CAS PubMed PubMed Central Google Scholar * Woese, C. R. On the evolution of cells. _Proc. Natl Acad. Sci. USA_ 99, 8742–8747
(2002). Article CAS PubMed PubMed Central Google Scholar * Vetsigian, K., Woese, C. R. & Goldenfeld, N. Collective evolution and the genetic code. _Proc. Natl Acad. Sci. USA_ 103,
10696–10701 (2006). A MAJOR ADVANCE IN CONSIDERING THE ROLE OF DARWINIAN VERSUS LAMARCKIAN INFLUENCES ON THE EVOLUTION OF THE GENETIC CODE, THE TRANSLATION PROCESS AND CELLULAR ORGANIZATION.
Article CAS PubMed PubMed Central Google Scholar * Woese, C. R., Olsen, G. J., Ibba, M. & Söll, D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process.
_Microbiol. Mol. Biol. Rev._ 64, 202–236 (2000). A COMPREHENSIVE REVIEW PLACING THE EVOLUTION OF AARSS IN THE CONTEXT OF THE REST OF THE TRANSLATION APPARATUS. Article CAS PubMed PubMed
Central Google Scholar * Woese, C. R. A new biology for a new century. _Microbiol. Mol. Biol. Rev._ 68, 173–186 (2004). A REFLECTION ON THE SCIENTIFIC PROCESS INVOLVED IN REVEALING THE
ARCHAEA TO THE SCIENTIFIC COMMUNITY. Article CAS PubMed PubMed Central Google Scholar * Kavran, J. M. et al. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code
innovation. _Proc. Natl Acad. Sci. USA_ 104, 11268–11273 (2007). Article CAS PubMed PubMed Central Google Scholar * Nozawa, K. et al. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure
reveals the molecular basis of orthogonality. _Nature_ 457, 1163–1167 (2009). Article CAS PubMed Google Scholar * Fournier, G. P., Huang, J. & Gogarten, J. P. Horizontal gene
transfer from extinct and extant lineages: biological innovation and the coral of life. _Philos. Trans. R. Soc. Lond. B. Biol. Sci._ 364, 2229–2239 (2009). A PROVOCATIVE VIEW OF THE
EVOLUTION OF LIFE (IN PARTICULAR, CONCERNING WHAT MIGHT BE INFERRED ABOUT EXTINCT LINEAGES). Article CAS PubMed PubMed Central Google Scholar * Pace, N. R. A molecular view of microbial
diversity and the biosphere. _Science_ 276, 734–740 (1997). AN INSIGHTFUL REVIEW CAPTURING THE EXCITEMENT EMERGING FROM THE USE OF DNA SEQUENCE DATA TO REVEAL THE EXTENT OF THE PREVIOUSLY
UNKNOWN PHYLOGENETIC AND FUNCTIONAL DIVERSITY IN THE MICROBIAL WORLD. Article CAS PubMed Google Scholar * Pace, N. R. Mapping the tree of life: progress and prospects. _Microbiol. Mol.
Biol. Rev._ 73, 565–576 (2009). AN UP-TO-DATE REVIEW ABOUT TREE CONSTRUCTION USING SSU RRNA SEQUENCES AND INFERENCES ABOUT THE UNIVERSAL TREE OF LIFE. Article CAS PubMed PubMed Central
Google Scholar * Huber, H. et al. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. _Nature_ 417, 63–67 (2002). INSIGHT INTO AN INTERESTING NEW LIFESTYLE AS
WELL AS A NEW DIVISION WITHIN THE ARCHAEA. Article CAS PubMed Google Scholar * Elkins, J. G. et al., A korarchaeal genome reveals insights into the evolution of the Archaea. _Proc. Natl
Acad. Sci. USA_ 105, 8102–8107 (2008). INSIGHT INTO AN INTERESTING NEW DIVISION WITHIN THE ARCHAEA. Article PubMed PubMed Central Google Scholar * Brochier-Armanet, C., Boussau, B.,
Gribaldo, S. & Forterre, P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. _Nature Rev. Microbiol._ 6, 245–252 (2008). Article CAS Google Scholar
* Casanueva, A. et al. Nanoarchaeal 16S rRNA gene sequences are widely dispersed in hyperthermophilic and mesophilic halophilic environments. _Extremophiles_ 12, 651–656 (2008). Article CAS
PubMed Google Scholar * Robertson, C. E., Harris, J. K., Spear, J. R. & Pace, N. R. Phylogenetic diversity and ecology of environmental Archaea. _Curr. Opin. Microbiol._ 8, 638–642
(2005). Article CAS PubMed Google Scholar * Ferry, J. G. Methanogenesis: Ecology, Physiology, Biochemistry and Genetics. _Chapman and Hall Microbiology Series_ (Chapman and Hall Inc.,
New York, 1993). A COMPENDIUM DESCRIBING ESSENTIALLY EVERYTHING THAT WAS KNOWN ABOUT METHANOGENS IN 1993. * Sohngen, N. L. Ueber bakterien, welche methan als kohlenstoffnahrung and
energiequelle gebrauchen. _Zentralbl. Bakteriol. Parasitik. Abt. I._ 15, 513–517 (1906) (in German). Google Scholar * Wolfe, R. S. The sixth A. J. Kluyver Memorial Lecture. Methanogens: a
surprising microbial group. _Antonie Van Leeuwenhoek_ 45, 353–364 (1979). Article CAS PubMed Google Scholar * Dalton, H. The Leeuwenhoek Lecture 2000: the natural and unnatural history
of methane-oxidizing bacteria. _Philos. Trans. R. Soc. Lond. B. Biol. Sci._ 360, 1207–1222 (2005). Article CAS PubMed PubMed Central Google Scholar * Barker, H. A. Studies upon the
methane-producing bacteria. _Arch. Mikrobiol._ 7, 420–438 (1936). Article CAS Google Scholar * Kluyver, A. J. & van Niel, C. B. Prospects for a natural system of classification of
bacteria. _Zentralbl. Bakterol. Parasitenkd. Infectinskr. Hyg. Abt. II_ 94, 369–403 (1936). Google Scholar * Barker, H. A. Studies upon the methane fermentation. IV. The isolation and
culture of _Methanobacterium omelianskii_. _Antonie van Leeuwenhoek J. Microbiol. Serol._ 6, 201–220 (1940). Article Google Scholar * Kandler, O. & Hippe, H. Lack of peptidoglycan in
the cell walls of _Methanosarcina barkeri_. _Arch. Microbiol._ 113, 57–60 (1977). Article CAS PubMed Google Scholar * Murray, P. A. & Zinder, S. Z. Nitrogen fixation by a
methanogenic archaebacterium. _Nature_ 312, 284–286 (1984). Article CAS Google Scholar * Belay, N., Sparkling, R. & Daniels, L. Dintrogen fixation by a thermophilic methanogenic
bacterium. _Nature_ 312, 286–288 (1984). Article CAS PubMed Google Scholar * DiMarco, A. A., Bobik, T. A. & Wolfe, R. S. Unusual coenzymes of methanogenesis. _Annu. Rev. Biochem._
59, 355–394 (1990). Article CAS PubMed Google Scholar * Ferry, J. G. How to make a living by exhaling methane. _Annu. Rev. Microbiol._ 64, 453–473 (2010). Article CAS PubMed Google
Scholar * Scheller, S., Goenrich, M., Boecher, R., Thauer, R. K. & Jaun, B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. _Nature_ 465, 606–608
(2010). Article CAS PubMed Google Scholar * Bult, C. A. et al. Complete genome sequence of the methanogenic archaeon, _Methanococcus jannaschii_. _Science_ 273, 1058–1073 (1996). A
HISTORICAL, EYE-OPENING ARTICLE DOCUMENTING THE FIRST GENOME SEQUENCE OF AN ARCHAEON, AND THE THIRD COMPLETE GENOME SEQUENCE OF AN ORGANISM. Article CAS PubMed Google Scholar * Orphan,
V. J., House, C. H., Hinrichs, K.-U., McKeegan, K. D. & DeLong, E. F. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. _Proc. Natl Acad. Sci. USA_ 99,
7663–7668 (2002). Article CAS PubMed PubMed Central Google Scholar * Knittel, K. & Boetius, A. Anaerobic oxidation of methane: progress with an unknown process. _Annu. Rev.
Microbiol._ 63, 311–334 (2009). A COMPREHENSIVE REVIEW OF ANAEROBIC METHANE OXIDATION. Article CAS PubMed Google Scholar * Alperin, M. & Hoehler, T. The ongoing mystery of sea-floor
methane. _Science_ 329, 288–289 (2010). Article CAS PubMed Google Scholar * Hinrichs, K. U., Hayes, J. M., Sylva, S. P., Brewer, P. G. & DeLong, E. F. Methane-consuming
archaebacteria in marine sediments. _Nature_ 398, 802–805 (1999). AN IMPORTANT BREAKTHROUGH IN REALIZING A ROLE FOR ARCHAEA IN ANAEROBIC METHANE OXIDATION. Article CAS PubMed Google
Scholar * Orphan, V. J., House, C. H., Hinrichs, K.-U., McKeegan, K. D. & DeLong, E. F. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis.
_Science_ 293, 484–487 (2001). Article CAS PubMed Google Scholar * Niemann, H. et al. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink.
_Nature_ 443, 854–858 (2006). Article CAS PubMed Google Scholar * Zehnder, A. J. & Brock, T. D. Methane formation and methane oxidation by methanogenic bacteria. _J. Bacteriol._ 137,
420–432 (1979). CAS PubMed PubMed Central Google Scholar * Hallam, S. J. et al. Reverse methanogenesis: testing the hypothesis with environmental genomics. _Science_ 305, 1457–1462
(2004). AN ILLUSTRATION OF HOW METAGENOMICS CAN BE USED TO TEASE OUT A PREVIOUSLY UNKNOWN PATHWAY WHEN THERE IS EVIDENCE THAT THE PROCESS (IN THIS CASE, ANAEROBIC OXIDATION OF METHANE)
OCCURS. Article CAS PubMed Google Scholar * Dekas, A. E., Poretsky, R. S. & Orphan, V. J. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. _Science_
326, 422–426 (2009). Article CAS PubMed Google Scholar * Michaelis, W. et al. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. _Science_ 297, 1013–1015 (2002).
Article CAS PubMed Google Scholar * Raghoebarsing, A. A. et al. A microbial consortium couples anaerobic methane oxidation to denitrification. _Nature_ 440, 918–921 (2006). Article CAS
PubMed Google Scholar * Ettwig, K. F. et al. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. _Environ. Microbiol._ 10, 3164–3173 (2008). Article CAS
PubMed Google Scholar * Ettwig, K. F. et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. _Nature_ 464, 543–548 (2010). A LESSON IN WHAT CAN BE LEARNED BEYOND THE
ARCHAEA, FROM THE ARCHAEA. Article CAS PubMed Google Scholar * Hao, B. et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. _Science_ 296, 1462–1466
(2002). AN IMPORTANT DESCRIPTION OF PYRROLYSINE AND THE MECHANISM OF ITS INCORPORATION INTO PROTEINS. Article CAS PubMed Google Scholar * Soares, J. A. et al. The residue mass of
L-pyrrolysine in three distinct methylamine methyltransferases. _J. Biol. Chem._ 280, 36962–36969 (2005). Article CAS PubMed Google Scholar * Krzycki, J. A. Function of genetically
encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. _Curr. Opin. Chem. Biol._ 8, 484–491 (2004). Article CAS PubMed Google Scholar * Heinemann, I. U. et al. The
appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. _Proc. Natl Acad. Sci. USA_ 106, 21103–21108 (2009). AN INTERESTING DESCRIPTION OF WHAT A NOVEL AMINO ACID AND
ITS COGNATE AARSS CAN EXPLAIN ABOUT EVOLUTION. Article PubMed PubMed Central Google Scholar * Farlow, W. G. in _United States Commission of Fish and Fisheries Part IV. Report for The
Commissioner 1878_ 969–973 (Government Printing Office, Washington DC, 1880). Google Scholar * Kocur, M. & Hodgkiss, W. Taxonomic status of the genus _Halococcus_ Schoop. _Int. J. Syst.
Bacteriol._ 23, 151–156 (1973). Article Google Scholar * Sehgal, S. N., Kates, M. & Gibbons, N. E. Lipids of _Halobacterium cutirubrum_. _Can. J. Biochem. Physiol._ 40, 69–81 (1962).
Article CAS PubMed Google Scholar * Marshall, C. L. & Brown, A. D. The membrane lipids of _Halobacterium halobium_. _Biochem. J._ 110, 441–448 (1968). Article CAS PubMed PubMed
Central Google Scholar * Fox, G. E. et al. The phylogeny of prokaryotes. _Science_ 209, 457–463 (1980). Article CAS PubMed Google Scholar * Oesterhelt, D. & Stoeckenius, W.
Rhodopsin-like protein from the purple membrane of _Halobacterium hatobium_. _Nature New Biol._ 233, 149–152 (1971). Article CAS PubMed Google Scholar * Oesterhelt, D. & Stoeckenius,
W. Functions of a new photoreceptor membrane. _Proc. Natl Acad. Sci. USA_ 70, 2853–2857 (1973). Article CAS PubMed PubMed Central Google Scholar * Henderson, R. & Unwin, P. N. T.
Three-dimensional model of purple membrane obtained by electron microscopy. _Nature_ 257, 28–32 (1975). AN IMPORTANT BREAKTHROUGH, NOT ONLY FOR RESEARCH INTO LIGHT-HARVESTING PROTEINS
(BACTERIORHODOPSIN), BUT ALSO FOR STRUCTURAL BIOLOGY STUDIES OF PROTEINS THAT CONTAIN MULTIPLE TRANSMEMBRANE DOMAINS. Article CAS PubMed Google Scholar * Spudich, J. L. & Bogomolni,
R. A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. _Nature_ 312, 509–513 (1984). Article CAS PubMed PubMed Central Google Scholar * Lanyi, J. K.
Bacteriorhodopsin as a model for proton pumps. _Nature_ 375, 461–463 (1995). Article CAS PubMed Google Scholar * Kolbe, M., Besir, H., Essen, L. O. & Oesterhelt, D. Structure of the
light-driven chloride pump halorhodopsin at 1.8 Å resolution. _Science_ 288, 1390–1396 (2000). Article CAS PubMed Google Scholar * Spudich, J. L., Yang, C. S., Jung, K. H. & Spudich,
E. N. Retinylidene proteins: structures and functions from Archaea to humans. _Annu. Rev. Cell. Dev. Biol._ 16, 365–392 (2000). Article CAS PubMed Google Scholar * Rasmussen, S. G. et
al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. _Nature_ 450, 383–387 (2007). Article CAS PubMed Google Scholar * Overington, J. P., Al-Lazikani, B. &
Hopkins, A. L. How many drug targets are there? _Nature Rev. Drug Discov._ 5, 993–936 (2006). Article CAS Google Scholar * Béjà, O. et al. Bacterial rhodopsin: evidence for a new type of
phototrophy in the sea. _Science_ 289, 1902–1906 (2000). AN IMPORTANT STUDY DESCRIBING A PREVIOUSLY UNKNOWN PHOTOTROPHIC SYSTEM FOR MARINE HETEROTROPHS. Article PubMed Google Scholar *
Béjà, O., Spudich, E. N., Spudich, J. L., Leclerc, M. & DeLong, E. F. Proteorhodopsin phototrophy in the ocean. _Nature_ 411, 786–789 (2001). Article PubMed Google Scholar * Venter,
J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. _Science_ 304, 66–74 (2004). Article PubMed Google Scholar * Frigaard, N. U., Martinez, A., Mincer, T. J. &
DeLong, E. F. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. _Nature_ 439, 847–850 (2006). Article CAS PubMed Google Scholar * Moran, M. A. &
Miller, W. L. Resourceful heterotrophs make the most of light in the coastal ocean. _Nature Rev. Microbiol._ 5, 792–800 (2007). Article CAS Google Scholar * Gómez-Consarnau, L. et al.
Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. _PLoS Biol._ 8, e1000358 (2010). Article CAS PubMed PubMed Central Google Scholar * DeLong, E. F.
& Béjà, O. The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. _PLoS Biol._ 8, e1000359 (2010). Article CAS PubMed PubMed Central Google
Scholar * Torsvik, T. & Dundas, I. D. Bacteriophage of _Halobacterium salinarium_. _Nature_ 248, 680–681 (1974). Article CAS PubMed Google Scholar * Dyall-Smith, M., Tang, S-L.
& Bath, C. Haloarchaeal viruses: how diverse are they? _Res. Microbiol._ 154, 309–313 (2003). Article PubMed Google Scholar * Prangishvili, D., Forterre, P. & Garrett, R. A.
Viruses of the Archaea: a unifying view. _Nature Rev. Microbiol._ 4, 837–848 (2006). Article CAS Google Scholar * Ortmann, A. C., Wiedenheft, B., Douglas, T. & Young, M. Hot
crenarchaeal viruses reveal deep evolutionary connections. _Nature Rev. Microbiol._ 4, 520–528 (2006). THIS ARTICLE, ALONG WITH REFERENCE 99, REVEALS THE DIVERSITY AND EXTENT OF ARCHAEAL
VIRUSES. Article CAS Google Scholar * Sapienza, C. & Doolittle, W. F. Unusual physical organization of the _Halobacterium_ genome. _Nature_ 295, 384–389 (1982). EARLY INSIGHT INTO
GENOME REARRANGEMENT THAT LED TO THEORIES ABOUT THE ROLE OF LGT IN EVOLUTION. Article CAS PubMed Google Scholar * Ng, W. V. et al. Genome sequence of _Halobacterium species_ NRC-1.
_Proc. Natl Acad. Sci. USA_ 97, 12176–12181 (2000). Article CAS PubMed PubMed Central Google Scholar * Papke, R. T., Koenig, J. E., Rodríguez-Valera, F. & Doolittle, W. F. Frequent
recombination in a saltern population of _Halorubrum_. _Science_ 306, 1928–1929 (2004). CAS PubMed Google Scholar * Papke, R. T. et al. Searching for species in haloarchaea. _Proc. Natl
Acad. Sci. USA_ 104, 14092–14097 (2007). Article CAS PubMed PubMed Central Google Scholar * Rodriguez-Valera, F. et al. Explaining microbial population genomics through phage predation.
_Nature Rev. Microbiol._ 7, 828–836 (2009). Article CAS Google Scholar * Baliga, N. S. et al. Coordinate regulation of energy transduction modules in _Halobacterium_ sp. analyzed by a
global systems approach. _Proc. Natl Acad. Sci. USA_ 99, 14913–14918 (2002). Article CAS PubMed PubMed Central Google Scholar * Facciotti, M. T. General transcription factor specified
global gene regulation in archaea _Proc. Natl Acad. Sci. USA_ 104, 4630–4635 (2007). Article CAS PubMed PubMed Central Google Scholar * Koide, T. et al. Prevalence of transcription
promoters within archaeal operons and coding sequences. _Mol. Syst. Biol._ 5, 285 (2009). Article CAS PubMed PubMed Central Google Scholar * Bonneau, R. et al. A predictive model for
transcriptional control of physiology in a free living cell. _Cell_ 131, 1354–1365 (2007). Article CAS PubMed Google Scholar * Bare, J. C., Koide, T., Reiss, D. J., Tenenbaum, D. &
Baliga, N. S. Integration and visualization of systems biology data in context of the genome. _BMC Bioinformatics_ 11, 382 (2010). Article CAS PubMed PubMed Central Google Scholar *
Rosenshine, I., Tchelet, R. & Mevarech, M. The mechanism of DNA transfer in the mating system of an archaebacterium. _Science_ 245, 1387–1389 (1989). Article CAS PubMed Google Scholar
* Humbard, M. A. et al. Ubiquitin-like small archaeal modifier proteins (SAMPs) in _Haloferax volcanii_. _Nature_ 463, 54–60 (2010). Article CAS PubMed PubMed Central Google Scholar *
Grogan, D. W. Exchange of genetic markers at extremely high temperatures in the archaeon _Sulfolobus acidocaldarius_. _J. Bacteriol._ 178, 3207–3211 (1996). Article CAS PubMed PubMed
Central Google Scholar * Bertani, G. & Baresi, L. Genetic transformation in the methanogen _Methanococcus voltae_ PS. _J. Bacteriol._ 169, 2730–2738 (1987). Article CAS PubMed
PubMed Central Google Scholar * Sato, T., Fukui, T., Atomi, H. & Imanaka, T. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon _Thermococcus
kodakaraensis_ KOD1. _J. Bacteriol._ 185, 210–220 (2003). Article CAS PubMed PubMed Central Google Scholar * Charlebois, R. L., Lam, W. L., Cline, S. W. & Doolittle, W. F.
Characterization of pHV2 from _Halobacterium volcanii_ and its use in demonstrating transformation of an archaebacterium. _Proc. Natl Acad. Sci. USA_ 84, 8530–8534 (1987). Article CAS
PubMed PubMed Central Google Scholar * Metcalf, W. W., Zhang, J. K., Apolinario, E., Sowers, K. R. & Wolfe, R. S. A genetic system for Archaea of the genus _Methanosarcina_:
liposome-mediated transformation and construction of shuttle vectors. _Proc. Natl Acad. Sci. USA_ 94, 2626–2631 (1997). A STRIKING EXAMPLE OF THE DEVELOPMENT OF A NOVEL, HIGH-EFFICIENCY GENE
TRANSFER SYSTEM. Article CAS PubMed PubMed Central Google Scholar * Schleper, C., Kubo, K. & Zillig, W. The particle SSV1 from the extremely thermophilic archaeon _Sulfolobus_ is a
virus: demonstration of infectivity and of transfection with viral DNA. _Proc. Natl Acad. Sci. USA_ 89, 7645–7649 (1992). Article CAS PubMed PubMed Central Google Scholar * Allers, T.
& Mevarech, M. Archaeal genetics — the third way. _Nature Rev. Genet._ 6, 58–73 (2005). A GOOD SUMMARY OF THE GENETIC SYSTEMS THAT HAVE BEEN DEVELOPED FOR ARCHAEA. Article CAS PubMed
Google Scholar * Sowers, K. & Anderson, K. in _Archaea: Molecular and Cellular Biology_ (ed. Cavicchioli, R.) 463–477 (American Society for Microbiology Press, Washington DC, 2007).
Book Google Scholar * Seghal, S. N., Kates, M. & Gibbons, N. E. Lipids of _Halobacterium cutirubrum_. _Can. J. Biochem. Physiol._ 40, 69–81 (1962). Article Google Scholar * Darland,
G., Brock, T. D., Samsonoff, W. & Conti, S. F. A thermophilic, acidophilic mycoplasma isolated from a coal refuse pile. _Science_ 170, 1416–1418 (1970). THE DISCOVERY OF AN UNUSUAL
ARCHAEON BEFORE THE KNOWLEDGE THAT A THIRD DOMAIN EXISTED. Article CAS PubMed Google Scholar * Langworthy, T. A., Smith, M. E. & Mayberry, W. R. A new class of lipopolysaccharide
from _Thermoplasma acidophilum_. _J. Bacteriol._ 119, 106–116 (1974). CAS PubMed PubMed Central Google Scholar * Brock, T. D., Brock, K. M., Belly, R. T. & Weiss, R. L. _Sulfolobus_:
a new genus of sulphur oxidizing bacteria living at low pH and high temperature. _Arch. Microbiol._ 84, 54–68 (1972). CAS Google Scholar * Langworthy, T. A., Smith, M. E. & Mayberry,
W. R. Long-chain glycerol diether and polyol dialkyl glycerol triether lipids of _Sulfolobus acidocaldarius_. _J. Bacteriol._ 112, 1193–1200 (1972). CAS PubMed PubMed Central Google
Scholar * Stetter, K. O. History of discovery of the first hyperthermophiles. _Extremophiles_ 10, 357–262 (2006). A BRIEF, BUT INFORMATIVE, REVIEW DESCRIBING HOW ARCHAEAL HYPERTHERMOPHILES
WERE DISCOVERED. Article PubMed Google Scholar * Stetter, K. O. Hyperthermophiles in the history of life. _Philos. Trans. R. Soc. Lond. B. Biol. Sci._ 361, 1837–1842 (2006). Article CAS
PubMed PubMed Central Google Scholar * Waters, E. et al. The genome of _Nanoarchaeum equitans_: insights into early archaeal evolution and derived parasitism. _Proc. Natl Acad. Sci.
USA_ 100, 12984–12988 (2003). Article CAS PubMed PubMed Central Google Scholar * Podar, M. et al. A genomic analysis of the archaeal system _Ignicoccus hospitalis-Nanoarchaeum
equitans_. _Genome_ _Biol._ 9, R158 (2008). Google Scholar * Küper, U., Meyer, C., Müller, V., Rachel, R. & Huber, H. Energized outer membrane and spatial separation of metabolic
processes in the hyperthermophilic Archaeon _Ignicoccus hospitalis_. _Proc. Natl Acad. Sci. USA_ 107, 3152–3126 (2010). INSIGHT INTO THE UNUSUAL RELATIONSHIP BETWEEN AN ARCHAEAL HOST AND
ARCHAEAL SYMBIONT OR PARASITE, WITH POSSIBLE IMPLICATIONS FOR THE EVOLUTION OF A EUKARYOTE-LIKE CELL TYPE. Article PubMed PubMed Central Google Scholar * van Ooij, C. A road to
eukaryotes? _Nature Rev. Microbiol._ 8, 246–247 (2010). Article CAS Google Scholar * Fuhrman, J. A., McCallum, K. & Davis, A. A. Novel major archaebacterial group from marine
plankton. _Nature_ 356, 148–149 (1992). Article CAS PubMed Google Scholar * DeLong, E. F. Archaea in coastal marine environments. _Proc. Natl Acad. Sci. USA_ 89, 5685–5689 (1992). ALONG
WITH REFERENCE 132, THIS PAPER DESCRIBES THE NEW INSIGHT THAT ARCHAEA ARE IMPORTANT MEMBERS OF MARINE ENVIRONMENTS. Article CAS PubMed PubMed Central Google Scholar * Karner, M. B.,
DeLong, E. F. & Karl, D. M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. _Nature_ 409, 507–510 (2001). THE SHEER ABUNDANCE OF ARCHAEA IN THE MARINE BIOSPHERE IS
DESCRIBED. Article CAS PubMed Google Scholar * Cavicchioli, R. Cold adapted Archaea. _Nature Rev. Microbiol._ 4, 331–343 (2006). INSIGHT INTO THE DIVERSITY, FUNCTIONAL ROLES AND ADAPTIVE
MECHANISMS OF ARCHAEA IN THE COLD BIOSPHERE. Article CAS Google Scholar * Franzmann, P. D. et al. _Halobacterium lacusprofundi_ sp. nov., a halophilic bacterium isolated from Deep Lake,
Antarctica. _Syst. Appl. Microbiol._ 11, 20–27 (1988). Article CAS Google Scholar * Franzmann, P. D., Stringer, N., Ludwig, W., Conway de Macario, E. & Rohde, M. A methanogenic
archaeon from Ace Lake, Antarctica: _Methanococcoides burtonii_ sp. nov. _System. Appl. Microbiol._ 15, 573–581 (1992). Article Google Scholar * Franzmann, P. D. et al. _Methanogenium
frigidum_ sp. nov., a psychrophilic, H2-using methanogen from Ace Lake, Antarctica. _Int. J. Syst. Bacteriol._ 47, 1068–1072 (1997). Article CAS PubMed Google Scholar * Allen, M. et al.
The genome sequence of the psychrophilic archaeon, _Methanococcoides burtonii_: the role of genome evolution in cold-adaptation. _ISME J._ 3, 1012–1035 (2009). Article CAS PubMed Google
Scholar * Takai, K. et al. Cell proliferation at 122 °C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. _Proc. Natl Acad. Sci. USA_
105, 10949–10954 (2008). Article PubMed PubMed Central Google Scholar * Schleper, C. et al. Genomic analysis reveals chromosomal variation in natural populations of the uncultured
psychrophilic archaeon _Cenarchaeum symbiosum_. _J. Bacteriol._ 180, 5003–5009 (1998). CAS PubMed PubMed Central Google Scholar * Schleper, C., Jurgens, G. & Jonuscheit, M. Genomic
studies of uncultivated archaea. _Nature Rev. Microbiol._ 3, 479–488 (2005). Article CAS Google Scholar * Könneke, M. et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon.
_Nature_ 437, 543–546 (2005). REVEALING INSIGHT INTO AN ELUSIVE PHYLOGENETIC AND PHENOTYPIC TYPE OF ARCHAEA: CHEMOLITHOAUTROPHIC AMMONIA OXIDIZERS. Article CAS PubMed Google Scholar *
Walker, C. B. et al. _Nitrosopumilus maritimus_ genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine _Crenarchaea_. _Proc. Natl Acad. Sci. USA_
107, 8818–8823 (2010). Article PubMed PubMed Central Google Scholar * Berg, I. A. et al. Autotrophic carbon fixation in archaea. _Nature Rev. Microbiol._ 8, 447–460 (2008). Article CAS
Google Scholar * Buckley, D. H., Graber, J. R. & Schmidt, T. M. Phylogenetic analysis of nonthermophilic members of the kingdom _Crenarchaeota_ and their diversity and abundance in
soils. _Appl. Environ. Microbiol._ 64, 4333–4339 (1998). CAS PubMed PubMed Central Google Scholar * Leininger, S. et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils.
_Nature_ 442, 806–809 (2006). Article CAS PubMed Google Scholar * Erguder, T. H., Boon, N., Wittebolle, L., Marzorati, M. & Verstraete, W. Environmental factors shaping the
ecological niches of ammonia-oxidizing archaea. _FEMS Microbiol. Rev._ 33, 855–869 (2009). Article CAS PubMed Google Scholar * Hirata, A., Klein, B. J. & Murakami, K. S. The X-ray
crystal structure of RNA polymerase from Archaea. _Nature_ 451, 851–854 (2008). A STRIKING EXAMPLE OF HOW VISUALLY AND FUNCTIONALLY SIMILAR KEY COMPONENTS OF THE INFORMATION PROCESSING
SYSTEMS ARE IN THE ARCHAEA AND EUKARYA. Article CAS PubMed PubMed Central Google Scholar * Cavicchioli, R., Siddiqui, K. S., Andrews, D. & Sowers, K. R. Low-temperature
extremophiles and their applications. _Curr. Opin. Biotech._ 13, 253–261 (2002). Article CAS PubMed Google Scholar * Schiraldi, C., Giuliano, M. & De Rosa, M. Perspectives on
biotechnological applications of archaea. _Archaea_ 1, 75–86 (2002). Article CAS PubMed PubMed Central Google Scholar * Auernik, K. S., Cooper, C. R. & Kelly, R. M. Life in hot
acid: pathway analyses in extremely thermoacidophilic archaea. _Curr. Opin. Biotechnol._ 19, 445–453 (2008). Article CAS PubMed PubMed Central Google Scholar * Krishnan, L. &
Sprott, G. D. Archaeosome adjuvants: immunological capabilities and mechanism(s) of action. _Vaccine_ 26, 2043–2055 (2008). Article CAS PubMed Google Scholar * Cavicchioli, R.
Extremophiles and the search for extra-terrestrial life. _Astrobiology_ 2, 281–292 (2002). Article CAS PubMed Google Scholar * Tung, H. C., Bramall, N. E. & Price, P. B. Microbial
origin of excess methane in glacial ice and implications for life on Mars. _Proc. Natl Acad. Sci. USA_ 102, 18292–18296 (2005). Article CAS PubMed PubMed Central Google Scholar *
Oremland, R. S. & Voytek, M. A. Acetylene as fast food: implications for development of life on anoxic primordial Earth and in the outer solar system. _Astrobiology_ 8, 45–58 (2008).
Article CAS PubMed Google Scholar * Oremland, R. S. NO connection with methane. _Nature_ 464, 500–501 (2010). Article CAS PubMed Google Scholar * Stetter, K. O. Hyperthermophilic
procaryotes. _FEMS Microbiol. Rev._ 18, 149–158 (1996). Article CAS Google Scholar * Koonin, E. V. & Martin, W. On the origin of genomes and cells within inorganic compartments.
_Trends Genet._ 21, 647–654 (2005). Article CAS PubMed PubMed Central Google Scholar * Brazelton, W. J. et al. Archaea and bacteria with surprising microdiversity show shifts in
dominance over 1,000-year time scales in hydrothermal chimneys. _Proc. Natl Acad. Sci. USA_ 107, 1612–1617 (2010). Article PubMed PubMed Central Google Scholar * Glansdorff, N., Xu, Y.
& Labedan, B. The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner. _Biol. Direct_ 3, 29 (2008). Article CAS PubMed PubMed Central
Google Scholar * (No authors listed). A sequence of changes. _Nature Rev. Microbiol._ 8, 85 (2010). * Delsuc, F., Brinkmann, H. & Philippe, H. Phylogenomics and the reconstruction of
the tree of life. _Nature Rev. Genet._ 6, 361–375 (2005). Article CAS PubMed Google Scholar * Doolittle, W. F. & Zhaxybayeva, O. On the origin of prokaryotic species. _Genome Res._
19, 744–756 (2009). Article CAS PubMed Google Scholar * Theobald, D. L. A formal test of the theory of universal common ancestry. _Nature_ 465, 219–222 (2010). Article CAS PubMed
Google Scholar * Pace, N. R. Time for a change. _Nature_ 441, 289 (2006). AN IMPORTANT AND AXIOMATIC ESSAY ABOUT LEARNING FROM WHAT WE HAVE LEARNED. Article CAS PubMed Google Scholar *
Pruesse, E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. _Nucleic Acids Res._ 35, 7188–7196 (2007). Article
CAS PubMed PubMed Central Google Scholar * Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. _Bioinformatics_ 22,
2688–2690 (2006). Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS I dedicate this Review to Carl Woese, who pioneered, led and continues to inspire the field with
his brilliance. I am indebted to C. Robertson, who generously constructed the phylogenetic tree, and M. DeMaere, who processed the RefSeq data. I also thank my many colleagues who informally
and formally commented on the manuscript, in particular N. Pace, T. Kolesnikow, F. Lauro, H. Ertan and M. Dyall-Smith. This Review could not be exhaustive and I regret not being able to
cite all of the relevant literature. Research in my laboratory is supported by the Australian Research Council. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * School of Biotechnology and
Biomolecular Sciences, The University of New South Wales, Sydney, 2052, NSW, Australia Ricardo Cavicchioli Authors * Ricardo Cavicchioli View author publications You can also search for this
author inPubMed Google Scholar ETHICS DECLARATIONS COMPETING INTERESTS The author declares no competing financial interests. RELATED LINKS RELATED LINKS FURTHER INFORMATION Ricardo
Cavicchioli's homepage NCBI Taxonomy Datbase PubMed RefSeq GLOSSARY * Domain The highest level of taxonomic division; the three domains are the Archaea, Bacteria and Eukarya. In
descending order, the other levels include: kingdom, phylum, class, order, family, genus and species. * Extremophile An organism that requires extreme environments for growth, such as
extremes of temperature, salinity or pH, or a combination of these. * Methanogen An anaerobic organism that generates methane by the reduction of carbon dioxide, acetic acid, or various
one-carbon compounds such as methylamines or methanol. * Halophile An organism that requires high concentrations of salt (typically greater than 1M NaCl) for growth. * Thermoacidophile An
organism that requires high temperatures (typically greater than 60 °C) and a low pH (typically less than pH 3) for growth. * Heterotroph An organism that uses organic compounds as nutrients
to produce energy for growth. * Phototrophic Pertaining to the growth of an organism: able to use sunlight to generate energy for growth. * Hyperthermophile An organism that requires
extremely high temperatures (typically greater than 80 °C) for growth. * Autotroph An organism that can grow on carbon dioxide as a sole source of carbon. * Small-subunit rRNA The ribosome
is the core biological machine of the translation apparatus and is essential for converting the genetic code described in DNA and mRNA into protein. Ribosomal RNA (rRNA) is the RNA component
of the ribosome and forms two subunits, the small subunit (SSU) and the large subunit. SSU rRNA is highly conserved in all cellular forms of life and is commonly used for describing the
phylogeny of organisms. * Lateral gene transfer Horizontal transfer of genes between unrelated species, as opposed to vertical inheritance within a species. * Bootstrap value A
computationally derived measure of confidence about tree topology: the closer the bootstrap value is to 100, the more confidence we can have in the topology of the tree. * Monophyletic
Pertaining to a natural taxonomic group or clade: consisting of individuals that share a common ancestor. * Reverse methanogenesis The methanogenesis pathway functioning in reverse to
consume methane and produce cellular carbon and energy; this process leads to the anaerobic oxidation of methane. * Chemolithoautotrophically Pertaining to an organism: able to derive energy
from a chemical reaction (chemotroph) using inorganic substrates as electron donors (lithotroph) and CO2 as a carbon source (autotroph). * Ammonia-oxidizing members of the Crenarchaeota An
archaeon with the ability to grow chemolithoautotrophically with near-stoichiometric conversion of ammonium cations (NH4+) to nitrite ions (NO2−) using carbonic acid (H2CO3) and ammonium
(NH4) as the sole sources of carbon and nitrogen, respectively. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Cavicchioli, R. Archaea — timeline of the
third domain. _Nat Rev Microbiol_ 9, 51–61 (2011). https://doi.org/10.1038/nrmicro2482 Download citation * Published: 06 December 2010 * Issue Date: January 2011 * DOI:
https://doi.org/10.1038/nrmicro2482 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative