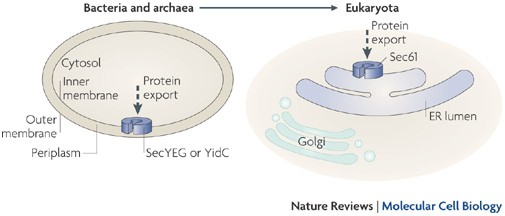
Delivering proteins for export from the cytosol
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Correct protein function depends on delivery to the appropriate cellular or subcellular location, and cells have evolved multiple strategies to achieve accurate protein targeting. The
delivery process necessitates that the protein either entirely crosses or is integrated into a distinct membrane.
In prokaryotes, protein export is defined as the delivery of the protein to the inner membrane or the periplasmic space. This process is conserved in eukaryotes and is exemplified by the
delivery of proteins to the membrane and lumen of the endoplasmic reticulum.
In post-translational delivery, proteins are delivered to the site of membrane translocation following their complete synthesis in a predominantly ATP-dependent process. Analogous mechanisms
for achieving this have been identified in both prokaryotes and eukaryotes, with a common theme being the use of molecular chaperones to maintain the protein in a partially unfolded state
suitable for membrane translocation.
Co-translational delivery is primarily mediated by the highly conserved signal recognition particle (SRP) and is dependent on GTP hydrolysis.
The interplay between these pathways seems to allow for a significant level of substrate specificity during protein targeting. Although the factors that determine the different steps of the
delivery process, and overlap between distinct delivery routes, are poorly understood, the resulting plasticity in protein delivery might allow for the modulation of cellular protein
delivery pathways in response to distinct environmental stresses.
Correct protein function depends on delivery to the appropriate cellular or subcellular compartment. Following the initiation of protein synthesis in the cytosol, many bacterial and
eukaryotic proteins must be integrated into or transported across a membrane to reach their site of function. Whereas in the post-translational delivery pathway ATP-dependent factors bind to
completed polypeptides and chaperone them until membrane translocation is initiated, a GTP-dependent co-translational pathway operates to couple ongoing protein synthesis to membrane
transport. These distinct pathways provide different solutions for the maintenance of proteins in a state that is competent for membrane translocation and their delivery for export from the
cytosol.
Our research is supported by funding from the Deutsche Forschungsgemeinschaft (DFG; SFB 638, GRK1188 and FOR 967) and the German–Israeli Foundation (I.S.), the Netherlands Organisation for
Scientific Research (NWO; J.L.) and the Biotechnology and Biological Sciences Research Council (BBSRC) and Wellcome Trust (B.C.S.C. and S.H.).
Faculty of Life Sciences, University of Manchester, Michael Smith Building, Oxford Road, Manchester, M13 9PT, UK
Biochemie-Zentrum der Universität Heidelberg (BZH), INF328, Heidelberg, 69120, Germany
Faculty of Earth and Life Sciences, VU University, de Boelelaan 1085, Amsterdam, 1081HV, The Netherlands
The site of convergence for protein export pathways, which is the inner membrane in prokaryotes and the endoplasmic reticulum membrane in eukaryotes.
A short span of amino acid residues that has no consensus sequence but is typically rich in hydrophobic amino acids and that provides the signal for protein export.
A cellular factor that binds to proteins and facilitates their folding, assembly and often export.
A domain in SecA that is formed by two helices of the helical scaffold domain.
A motif containing two sequential Arg residues. This motif defines the signal sequence to direct substrates to the twin Arg translocon.
A Met-rich domain of the signal recognition particle SRP54 that both houses the signal sequence recognition region and binds the SRP RNA.
A domain in the signal recognition particle SRP54 that comprises a four helix bundle and the GTPase domain.
The soluble region in the chloroplast that is functionally synonymous with bacterial cytosol.
A domain of the signal recognition particle (SRP) that comprises SRP9 and SRP14, together with helices three and four of the SRP RNA.
Anyone you share the following link with will be able to read this content: