Mutational effects and the evolution of new protein functions
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
KEY POINTS * The divergence of new genes and proteins occurs through mutations that modulate protein function. The effects of these mutations are pleiotropic, thus imposing trade-offs
between selection pressures for the existing function and the newly evolving one and among the protein's activity, stability and dosage. * Various compensatory and buffering mechanisms,
such as gene duplication, upregulation of expression, stabilizing mutations and chaperone folding assistance, can alleviate these trade-offs and so facilitate functional divergence. *
Despite buffering effects, the fitness distribution of mutations at the protein level, and for whole organisms, is such that most of the mutations are either neutral or deleterious. This
results in the rapid and irreversible non-functionalization of proteins that accumulate mutations under no selection. * The distribution of fitness effects of mutations for whole organisms
is comparable, and possibly even more deleterious, than that of protein mutations. * Duplication underlies the divergence of new genes and proteins. Duplication is almost as frequent as
point mutations and is a common mechanism for resolving the trade-off conflicts that arise owing to parallel selection pressures. These pressures may regard the existing and the new function
and maintenance of the protein's structural stability. * Duplication, and the emergence of new genes and proteins, may occur at different stages of the divergence process. The
selection pressures that act on the gene and its duplicate may differ, giving rise to different mechanisms of divergence. These mechanisms are described under three schematic models —
Ohno's model, the 'divergence before duplication' (DPD) model and the sub-functionalization model. * In Ohno's model of divergence, duplication is a neutral event. The
duplicated copy of the protein drifts under no selection until a new function becomes under selection. The downside of this model is that under no selection, non-functionalization of the
drifting protein is inevitable. Its advantage is that divergence is independent of trade-offs between the new and existing functions. * The DPD model is based on a 'generalist'
intermediate that confers a selectable degree of both the new and existing functions. Duplication occurs after the acquisition of a new function, and occurs under positive selection to
increase protein dosage and/or alleviate trade-offs that make the acquisition of new function depend on loss of the existing one. * The sub-functionalization model combines elements of the
DPD model and Ohno's model. Duplication is initially a neutral event, but once mutations that partially reduce protein activity or dosage appear, both copies must remain functional.
Duplication therefore enables a larger genetic variability to accumulate, thereby facilitating the emergence of new functions. * The DPD and sub-functionalization models are both based on
mutations with adaptive potential initially accumulating as neutral. As such, they are related to the notions of hidden or apparently neutral variation and of neutral networks. ABSTRACT The
divergence of new genes and proteins occurs through mutations that modulate protein function. However, mutations are pleiotropic and can have different effects on organismal fitness
depending on the environment, as well as opposite effects on protein function and dosage. We review the pleiotropic effects of mutations. We discuss how they affect the evolution of gene and
protein function, and how these complex mutational effects dictate the likelihood and mechanism of gene duplication and divergence. We propose several factors that can affect the divergence
of new protein functions, including mutational trade-offs and hidden, or apparently neutral, variation. Access through your institution Buy or subscribe This is a preview of subscription
content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue
Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL
ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS FUNCTIONAL SYNONYMOUS MUTATIONS AND THEIR
EVOLUTIONARY CONSEQUENCES Article 20 May 2025 THE POPULATION GENOMICS OF ADAPTIVE LOSS OF FUNCTION Article Open access 11 February 2021 CHANGES IN THE DISTRIBUTION OF FITNESS EFFECTS AND
ADAPTIVE MUTATIONAL SPECTRA FOLLOWING A SINGLE FIRST STEP TOWARDS ADAPTATION Article Open access 31 August 2021 REFERENCES * Conant, G. C. & Wolfe, K. H. Turning a hobby into a job: how
duplicated genes find new functions. _Nature Rev. Genet._ 9, 938–950 (2008). CAS PubMed Google Scholar * Innan, H. & Kondrashov, F. The evolution of gene duplications: classifying and
distinguishing between models. _Nature Rev. Genet._ 11, 97–108 (2010). CAS PubMed Google Scholar * DePristo, M. A., Weinreich, D. M. & Hartl, D. L. Missense meanderings in sequence
space: a biophysical view of protein evolution. _Nature Rev. Genet._ 6, 678–687 (2005). CAS PubMed Google Scholar * Pal, C., Papp, B. & Lercher, M. J. An integrated view of protein
evolution. _Nature Rev. Genet._ 7, 337–348 (2006). CAS PubMed Google Scholar * Dean, A. M. & Thornton, J. W. Mechanistic approaches to the study of evolution: the functional
synthesis. _Nature Rev. Genet._ 8, 675–688 (2007). CAS PubMed Google Scholar * Tokuriki, N. & Tawfik, D. S. Stability effects of mutations and protein evolvability. _Curr. Opin.
Struct. Biol._ 19, 596–604 (2009). CAS PubMed Google Scholar * Eyre-Walker, A. & Keightley, P. D. The distribution of fitness effects of new mutations. _Nature Rev. Genet._ 8, 610–618
(2007). CAS PubMed Google Scholar * Camps, M., Herman, A., Loh, E. & Loeb, L. A. Genetic constraints on protein evolution. _Crit. Rev. Biochem. Mol. Biol._ 42, 313–326 (2007). CAS
PubMed Google Scholar * Bloom, J. D. et al. Thermodynamic prediction of protein neutrality. _Proc. Natl Acad. Sci. USA_ 102, 606–611 (2005). CAS PubMed PubMed Central Google Scholar *
Bershtein, S., Segal, M., Bekerman, R., Tokuriki, N. & Tawfik, D. S. Robustness–epistasis link shapes the fitness landscape of a randomly drifting protein. _Nature_ 444, 929–932 (2006).
CAS PubMed Google Scholar * Bershtein, S. & Tawfik, D. S. Ohno's model revisited: measuring the frequency of potentially adaptive mutations under various mutational drifts. _Mol.
Biol. Evol._ 25, 2311–2318 (2008). CAS PubMed Google Scholar * Hecky, J. & Muller, K. M. Structural perturbation and compensation by directed evolution at physiological temperature
leads to thermostabilization of β-lactamase. _Biochemistry_ 44, 12640–12654 (2005). CAS PubMed Google Scholar * Yue, P. & Moult, J. Identification and analysis of deleterious human
SNPs. _J. Mol. Biol._ 356, 1263–1274 (2006). CAS PubMed Google Scholar * Tokuriki, N., Oldfield, C. J., Uversky, V. N., Berezovsky, I. N. & Tawfik, D. S. Do viral proteins possess
unique biophysical features? _Trends Biochem. Sci._ 34, 53–59 (2009). CAS PubMed Google Scholar * Wang, X., Minasov, G. & Shoichet, B. K. Evolution of an antibiotic resistance enzyme
constrained by stability and activity trade-offs. _J. Mol. Biol._ 320, 85–95 (2002). CAS PubMed Google Scholar * Tokuriki, N., Stricher, F., Serrano, L. & Tawfik, D. S. How protein
stability and new functions trade off. _PLoS Comput. Biol._ 4, e1000002 (2008). PubMed PubMed Central Google Scholar * Levin, K. B. et al. Following evolutionary paths to protein–protein
interactions with high affinity and selectivity. _Nature Struct. Mol. Biol._ 16, 1049–1055 (2009). CAS Google Scholar * Lindner, A. B., Madden, R., Demarez, A., Stewart, E. J. &
Taddei, F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. _Proc. Natl Acad. Sci. USA_ 105, 3076–3081 (2008). CAS PubMed PubMed Central
Google Scholar * McLoughlin, S. Y. & Copley, S. D. A compromise required by gene sharing enables survival: implications for evolution of new enzyme activities. _Proc. Natl Acad. Sci.
USA_ 105, 13497–13502 (2008). CAS PubMed PubMed Central Google Scholar * Vick, J. E., Schmidt, D. M. & Gerlt, J. A. Evolutionary potential of (β/α)8-barrels: _in vitro_ enhancement
of a 'new' reaction in the enolase superfamily. _Biochemistry_ 44, 11722–11729 (2005). CAS PubMed Google Scholar * Khersonsky, O. & Tawfik, D. S. Enzyme promiscuity: a
mechanistic and evolurtionary perpective. _Ann. Rev. Biochem._ 79, 471–505 (2010). CAS PubMed Google Scholar * Aharoni, A. et al. The 'evolvability' of promiscuous protein
functions. _Nature Genet._ 37, 73–76 (2005). CAS PubMed Google Scholar * Tokuriki, N. & Tawfik, D. S. Protein dynamism and evolvability. _Science_ 324, 203–207 (2009). CAS PubMed
Google Scholar * Scannell, D. R. & Wolfe, K. H. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. _Genome Res._ 18,
137–147 (2008). CAS PubMed PubMed Central Google Scholar * Kaessmann, H. Genetics. More than just a copy. _Science_ 325, 958–959 (2009). PubMed Google Scholar * Parker, H. G. et al. An
expressed _fgf4_ retrogene is associated with breed-defining chondrodysplasia in domestic dogs. _Science_ 325, 995–998 (2009). CAS PubMed PubMed Central Google Scholar * Andersson, D.
I. & Hughes, D. Gene amplification and adaptive evolution in bacteria. _Annu. Rev. Genet._ 43, 167–195 (2009). CAS PubMed Google Scholar * Schimke, R. T. Gene amplification in
cultured cells. _J. Biol. Chem._ 263, 5989–5992 (1988). CAS PubMed Google Scholar * Papp, B., Pal, C. & Hurst, L. D. Metabolic network analysis of the causes and evolution of enzyme
dispensability in yeast. _Nature_ 429, 661–664 (2004). CAS PubMed Google Scholar * Perry, G. H. et al. Diet and the evolution of human amylase gene copy number variation. _Nature Genet._
39, 1256–1260 (2007). CAS PubMed Google Scholar * Fablet, M., Bueno, M., Potrzebowski, L. & Kaessmann, H. Evolutionary origin and functions of retrogene introns. _Mol. Biol. Evol._
26, 2147–2156 (2009). CAS PubMed Google Scholar * Jablonka, E. & Lamb, M. J. _Epigenetic Inheritance and Evolution: The Lamarckian Dimension_ (Oxford Univ. Press, Oxford, UK, 1995).
Google Scholar * Steele, E. J., Lindley, R. A. & Blanden, R. V. _Lamarck's Signature: How Retrogenes Are Changing Darwin's Natural Selection Paradigm_, (Allen & Unwin;
Perseus Books, Australia, 1988). Google Scholar * Chen, G. K. et al. Preferential expression of a mutant allele of the amplified _MDR1_ (_ABCB1_) gene in drug-resistant variants of a human
sarcoma. _Genes Chromosomes Cancer_ 34, 372–383 (2002). PubMed Google Scholar * Qian, W. & Zhang, J. Gene dosage and gene duplicability. _Genetics_ 179, 2319–2324 (2008). PubMed
PubMed Central Google Scholar * Goldsmith, M. & Tawfik, D. S. Potential role of phenotypic mutations in the evolution of protein expression and stability. _Proc. Natl Acad. Sci. USA_
106, 6197–6202 (2009). CAS PubMed PubMed Central Google Scholar * Siu, L. K., Ho, P. L., Yuen, K. Y., Wong, S. S. & Chau, P. Y. Transferable hyperproduction of TEM-1 β-lactamase in
_Shigella flexneri_ due to a point mutation in the pribnow box. _Antimicrob. Agents Chemother._ 41, 468–470 (1997). CAS PubMed PubMed Central Google Scholar * Hall, B. G. Evolution of a
regulated operon in the laboratory. _Genetics_ 101, 335–344 (1982). CAS PubMed PubMed Central Google Scholar * Hall, B. G. The EBG system of _E. coli_: origin and evolution of a novel
β-galactosidase for the metabolism of lactose. _Genetica_ 118, 143–156 (2003). CAS PubMed Google Scholar * Stoebel, D. M., Dean, A. M. & Dykhuizen, D. E. The cost of expression of
_Escherichia coli_ lac operon proteins is in the process, not in the products. _Genetics_ 178, 1653–1660 (2008). CAS PubMed PubMed Central Google Scholar * Wagner, A. Energy constraints
on the evolution of gene expression. _Mol. Biol. Evol._ 22, 1365–1374 (2005). CAS PubMed Google Scholar * Vavouri, T., Semple, J. I., Garcia-Verdugo, R. & Lehner, B. Intrinsic protein
disorder and interaction promiscuity are widely associated with dosage sensitivity. _Cell_ 138, 198–208 (2009). CAS PubMed Google Scholar * Veitia, R. A. Gene dosage balance: deletions,
duplications and dominance. _Trends Genet._ 21, 33–35 (2005). CAS PubMed Google Scholar * Drummond, D. A., Bloom, J. D., Adami, C., Wilke, C. O. & Arnold, F. H. Why highly expressed
proteins evolve slowly. _Proc. Natl Acad. Sci. USA_ 102, 14338–14343 (2005). CAS PubMed PubMed Central Google Scholar * Fares, M. A., Ruiz- González, M. X., Moya, A., Elena, S. F. &
Barrio, E. Endosymbiotic bacteria: GroEL buffers against deleterious mutations. _Nature_ 417, 398 (2002). CAS PubMed Google Scholar * Rutherford, S., Hirate, Y. & Swalla, B. J. The
Hsp90 capacitor, developmental remodeling, and evolution: the robustness of gene networks and the curious evolvability of metamorphosis. _Crit. Rev. Biochem. Mol. Biol._ 42, 355–372 (2007).
CAS PubMed Google Scholar * Cowen, L. E. & Lindquist, S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. _Science_ 309, 2185–2189 (2005). CAS
PubMed Google Scholar * Parent, K. N., Ranaghan, M. J. & Teschke, C. M. A second-site suppressor of a folding defect functions via interactions with a chaperone network to improve
folding and assembly _in vivo_. _Mol. Microbiol._ 54, 1036–1050 (2004). CAS PubMed Google Scholar * Tokuriki, N. & Tawfik, D. S. Chaperonin overexpression promotes genetic variation
and enzyme evolution. _Nature_ 459, 668–673 (2009). CAS PubMed Google Scholar * Zhang, L. & Watson, L. T. Analysis of the fitness effect of compensatory mutations. _HFSP J._ 3, 47–54
(2009). PubMed Google Scholar * Bershtein, S., Goldin, K. & Tawfik, D. S. Intense neutral drifts yield robust and evolvable consensus proteins. _J. Mol. Biol._ 379, 1029–1044 (2008).
CAS PubMed Google Scholar * Hecky, J., Mason, J. M., Arndt, K. M. & Muller, K. M. A general method of terminal truncation, evolution, and re-elongation to generate enzymes of enhanced
stability. _Methods Mol. Biol._ 352, 275–304 (2007). CAS PubMed Google Scholar * Kather, I., Jakob, R. P., Dobbek, H. & Schmid, F. X. Increased folding stability of TEM-1 β-lactamase
by _in vitro_ selection. _J. Mol. Biol._ 383, 238–251 (2008). CAS PubMed Google Scholar * Marciano, D. C. et al. Genetic and structural characterization of an L201P global suppressor
substitution in TEM-1 β-lactamase. _J. Mol. Biol._ 384, 151–164 (2008). CAS PubMed PubMed Central Google Scholar * Kimura, M. The role of compensatory neutral mutations in molecular
evolution. _J. Genet._ 64, 7–19 (1985). CAS Google Scholar * Bloom, J. D., Labthavikul, S. T., Otey, C. R. & Arnold, F. H. Protein stability promotes evolvability. _Proc. Natl Acad.
Sci. USA_ 103, 5869–5874 (2006). CAS PubMed PubMed Central Google Scholar * McIntosh, B. E., Hogenesch, J. B. & Bradfield, C. A. Mammalian Per-Arnt-Sim proteins in environmental
adaptation. _Annu. Rev. Physiol._ 72, 625–645 (2010). CAS PubMed Google Scholar * Lynch, M. Genomics. Gene duplication and evolution. _Science_ 297, 945–947 (2002). CAS PubMed Google
Scholar * Beckmann, J. S., Estivill, X. & Antonarakis, S. E. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. _Nature Rev.
Genet._ 8, 639–646 (2007). CAS PubMed Google Scholar * Hastings, P. J., Lupski, J. R., Rosenberg, S. M. & Ira, G. Mechanisms of change in gene copy number. _Nature Rev. Genet._ 10,
551–564 (2009). CAS PubMed Google Scholar * Liao, B. Y. & Zhang, J. Null mutations in human and mouse orthologs frequently result in different phenotypes. _Proc. Natl Acad. Sci. USA_
105, 6987–6992 (2008). CAS PubMed PubMed Central Google Scholar * Ohno, S. _Evolution by Gene Duplication_ (Allen & Unwin; Springer, New York, 1970). Google Scholar * Kimura, M.
& Ota, T. On some principles governing molecular evolution. _Proc. Natl Acad. Sci. USA_ 71, 2848–2852 (1974). CAS PubMed PubMed Central Google Scholar * Zhang, J. Evolution by gene
duplication: an update. _Trends Ecol. Evol._ 18, 292–298 (2003). Google Scholar * Hughes, A. L. Adaptive evolution after gene duplication. _Trends Genet._ 18, 433–434 (2002). CAS PubMed
Google Scholar * Lynch, M. & Katju, V. The altered evolutionary trajectories of gene duplicates. _Trends Genet._ 20, 544–549 (2004). CAS PubMed Google Scholar * Kondrashov, F. A.
& Koonin, E. V. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. _Trends Genet._ 20, 287–290 (2004). CAS PubMed Google
Scholar * Bergthorsson, U., Andersson, D. I. & Roth, J. R. Ohno's dilemma: evolution of new genes under continuous selection. _Proc. Natl Acad. Sci. USA_ 104, 17004–17009 (2007).
CAS PubMed PubMed Central Google Scholar * Kondrashov, F. A. In search of the limits of evolution. _Nature Genet._ 37, 9–10 (2005). CAS PubMed Google Scholar * Boehr, D. D., Nussinov,
R. & Wright, P. E. The role of dynamic conformational ensembles in biomolecular recognition. _Nature Chem. Biol._ 5, 789–796 (2009). CAS Google Scholar * Piatigorsky, J. et al. Gene
sharing by D-crystallin and argininosuccinate lyase. _Proc. Natl Acad. Sci. USA_ 85, 3479–3483 (1988). CAS PubMed PubMed Central Google Scholar * Piatigorsky, J. _Gene Sharing and
Evolution: The Diversity of Protein Functions_, (Harvard Univ. Press, Cambridge, Massachusetts, USA; London, UK, 2007). Google Scholar * Lee, Y. N., Nechushtan, H., Figov, N. & Razin,
E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FceRI-activated mast cells. _Immunity_ 20, 145–151 (2004). CAS PubMed Google Scholar *
Sedlak, T. W. & Snyder, S. H. Messenger molecules and cell death: therapeutic implications. _JAMA_ 295, 81–89 (2006). CAS PubMed Google Scholar * Rosenberg, H. F. RNase A
ribonucleases and host defense: an evolving story. _J. Leukoc. Biol._ 83, 1079–1087 (2008). CAS PubMed Google Scholar * Jensen, R. A. Enzyme recruitment in evolution of new function.
_Annu. Rev. Microbiol._ 30, 409–425 (1974). Google Scholar * O'Brien, P. J. & Herschlag, D. Catalytic promiscuity and the evolution of new enzymatic activities. _Chem. Biol._ 6,
R91–R105 (1999). CAS PubMed Google Scholar * Palmer, D. R. et al. Unexpected divergence of enzyme function and sequence: '_N_-acylamino acid racemase' is _o_-succinylbenzoate
synthase. _Biochemistry_ 38, 4252–4258 (1999). CAS PubMed Google Scholar * James, L. C. & Tawfik, D. S. Catalytic and binding poly-reactivities shared by two unrelated proteins: the
potential role of promiscuity in enzyme evolution. _Protein Sci._ 10, 2600–2607 (2001). CAS PubMed PubMed Central Google Scholar * Afriat, L., Roodveldt, C., Manco, G. & Tawfik, D.
S. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. _Biochemistry_ 45, 13677–13686 (2006). CAS PubMed Google Scholar *
Copley, S. D. Evolution of efficient pathways for degradation of anthropogenic chemicals. _Nature Chem. Biol._ 5, 559–566 (2009). CAS Google Scholar * Copley, S. D. _Comprehensive Natural
Products II: Chemistry and Biology_ (eds Mander, L. & Liu, H.-W.) (Elsevier, Oxford, 2010). Google Scholar * Hughes, A. L. The evolution of functionally novel proteins after gene
duplication. _Proc. Biol. Sci._ 256, 119–124 (1994). CAS PubMed Google Scholar * Barkman, T. & Zhang, J. Evidence for escape from adaptive conflict? _Nature_ 462, e1; discussion e2–e3
(2009). CAS PubMed Google Scholar * Des Marais, D. L. & Rausher, M. D. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. _Nature_ 454, 762–765 (2008).
CAS PubMed Google Scholar * Lynch, M. & Force, A. The probability of duplicate gene preservation by subfunctionalization. _Genetics_ 154, 459–473 (2000). CAS PubMed PubMed Central
Google Scholar * Dykhuizen, D. & Hartl, D. L. Selective neutrality of 6PGD allozymes in _E. coli_ and the effects of genetic background. _Genetics_ 96, 801–817 (1980). CAS PubMed
PubMed Central Google Scholar * Force, A. et al. Preservation of duplicate genes by complementary, degenerative mutations. _Genetics_ 151, 1531–1545 (1999). CAS PubMed PubMed Central
Google Scholar * Nei, M. The new mutation theory of phenotypic evolution. _Proc. Natl Acad. Sci. USA_ 104, 12235–12242 (2007). CAS PubMed PubMed Central Google Scholar * Wagner, A.
_Robustness and Evolvability in Living Systems_ (Princeton Univ. Press, Princeton, USA, 2005). Google Scholar * Schuster, P. & Fontana, W. Chance and necessity in evolution: lessons
from RNA. _Physica D_ 133, 427–452 (1999). CAS Google Scholar * Wroe, R., Chan, H. S. & Bornberg-Bauer, E. A structural model of latent evolutionary potentials underlying neutral
networks in proteins. _HFSP J._ 1, 79–87 (2007). CAS PubMed PubMed Central Google Scholar * Klassen, J. L. Pathway evolution by horizontal transfer and positive selection is accommodated
by relaxed negative selection upon upstream pathway genes in purple bacterial carotenoid biosynthesis. _J. Bacteriol._ 191, 7500–7508 (2009). CAS PubMed PubMed Central Google Scholar *
Wloch, D. M., Szafraniec, K., Borts, R. H. & Korona, R. Direct estimate of the mutation rate and the distribution of fitness effects in the yeast _Saccharomyces cerevisiae_. _Genetics_
159, 441–452 (2001). CAS PubMed PubMed Central Google Scholar * Kivisaar, M. Degradation of nitroaromatic compounds: a model to study evolution of metabolic pathways. _Mol. Microbiol._
74, 777–781 (2009). CAS PubMed Google Scholar * Wackett, L. P. Questioning our perceptions about evolution of biodegradative enzymes. _Curr. Opin. Microbiol._ 12, 244–251 (2009). CAS
PubMed Google Scholar * Newcomb, R. D., Gleeson, D. M., Yong, C. G., Russell, R. J. & Oakeshott, J. G. Multiple mutations and gene duplications conferring organophosphorus insecticide
resistance have been selected at the Rop-1 locus of the sheep blowfly, _Lucilia cuprina_. _J. Mol. Evol._ 60, 207–220 (2005). CAS PubMed Google Scholar * Patzoldt, W. L., Hager, A. G.,
McCormick, J. S. & Tranel, P. J. A codon deletion confers resistance to herbicides inhibiting protoporphyrinogen oxidase. _Proc. Natl Acad. Sci. USA_ 103, 12329–12334 (2006). CAS PubMed
PubMed Central Google Scholar * O'Maille, P. E. et al. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. _Nature Chem. Biol._
4, 617–623 (2008). CAS Google Scholar * Lozovsky, E. R. et al. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. _Proc. Natl Acad. Sci. USA_ 106, 12025–12030
(2009). CAS PubMed PubMed Central Google Scholar * Poelwijk, F. J., Kiviet, D. J., Weinreich, D. M. & Tans, S. J. Empirical fitness landscapes reveal accessible evolutionary paths.
_Nature_ 445, 383–386 (2007). CAS PubMed Google Scholar * Kondrashov, A. S., Sunyaev, S. & Kondrashov, F. A. Dobzhansky–Muller incompatibilities in protein evolution. _Proc. Natl
Acad. Sci. USA_ 99, 14878–14883 (2002). CAS PubMed PubMed Central Google Scholar * Weinreich, D. M., Delaney, N. F., Depristo, M. A. & Hartl, D. L. Darwinian evolution can follow
only very few mutational paths to fitter proteins. _Science_ 312, 111–114 (2006). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS D.S.T. is the incumbent of the Nella and
Leon Benoziyo Professorial Chair. Financial support from the Meil de Botton Aynsley and the EU network BioModularH2 are gratefully acknowledged. We are very grateful to S. Bershtein, N.
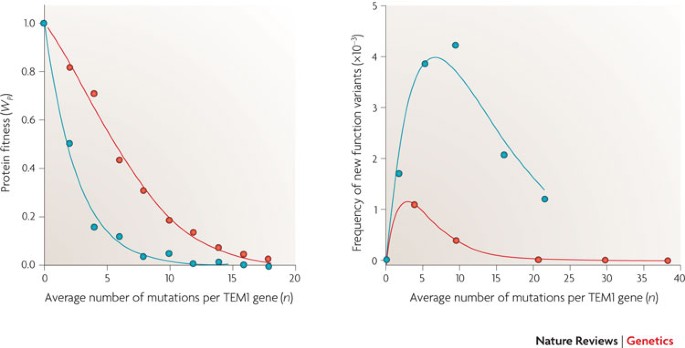
Tokuriki, F. Kondrashov and J. G. Zhang for their insightful comments regarding this manuscript and to A. Eyre-Walker for providing the data for the figure in Box 1. AUTHOR INFORMATION
AUTHORS AND AFFILIATIONS * Department of Biological Chemistry, Weizmann Institute of Science, Rehovot, 76100, Israel Misha Soskine & Dan S. Tawfik Authors * Misha Soskine View author
publications You can also search for this author inPubMed Google Scholar * Dan S. Tawfik View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING
AUTHOR Correspondence to Dan S. Tawfik. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. GLOSSARY * Protein mutations Missense mutations that
occur in encoded open reading frames. * Trade-offs Gains of a new activity or property at the expense of other activities or properties. * Protein stability The capacity of a protein to
adopt its native, functional structure. Stability also correlates with cellular protein levels. * Sub-functionalization Degenerate mutations that result in a gene and its duplicated copy
sharing the burden of one function. * Negative epistasis The combined effect of mutations being more deleterious than expected from their individual effects. * Protein fitness Levels of
physiological function exerted by a given protein variant under a certain selection pressure. * Non-functionalization The complete inactivation of a gene or protein by highly deleterious
mutations. * Neo-functionalization The divergence of a duplicated gene or protein to execute a new function. * ΔΔG The stability difference for a protein variant versus its wild-type
reference (ΔΔG > 0 indicates lower stability). * Disordered domains Protein domains with a high degree of random coil and loop regions and a low degree of highly ordered secondary
structure. * Apparently neutral mutations Mutations that have no significant or observable fitness effect under a given environment. * New-function mutations Mutations that mediate changes
in protein activity, typically by increasing a weak, latent promiscuous function. * New–existing function trade-offs The acquisition of a new function through mutations that undermine the
existing function. * Chaperones Proteins that mediate the correct folding and assembly of other proteins. * Specialists Genes or proteins that exert one specific function with high
proficiency. * Generalist A gene or protein that exerts multiple functions, typically one primary function and additional secondary or promiscuous functions. * New-function–stability
trade-offs Mutations that increase the new, evolving function but reduce protein stability and protein dosage. * Productive variation Genetic variation that does not compromise fitness in
the dwelling environment but holds the potential for adaptation to new environments. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Soskine, M., Tawfik,
D. Mutational effects and the evolution of new protein functions. _Nat Rev Genet_ 11, 572–582 (2010). https://doi.org/10.1038/nrg2808 Download citation * Issue Date: August 2010 * DOI:
https://doi.org/10.1038/nrg2808 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative