Arrayed cellular microenvironments for identifying culture and differentiation conditions for stem, primary and rare cell populations
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
During the development of an organism, cells are exposed to a myriad of signals, structural components and scaffolds, which collectively make up the cellular microenvironment. The majority
of current developmental biology studies examine the effect of individual or small subsets of molecules and parameters on cellular behavior, and they consequently fail to explore the
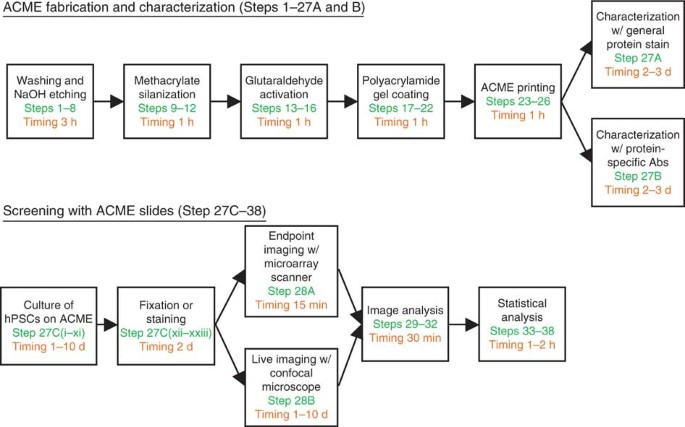
complexity of factors to which cells are exposed. Here we describe a technology, referred to as arrayed cellular microenvironments (ACMEs), that allows for a high-throughput examination of
the effects of multiple extracellular components in a combinatorial manner on any cell type of interest. We will specifically focus on the application of this technology to human pluripotent
stem cells (hPSCs), a population of cells with tremendous therapeutic potential, and one for which growth and differentiation conditions are poorly characterized and far from defined and
optimized. A standard ACME screen uses the technologies previously applied to the manufacture and analysis of DNA microarrays, requires standard cell-culture facilities and can be performed
from beginning to end within 5–10 days.
D.A.B. was supported by funding from the University of California San Diego Stem Cell Program and by a gift from Michael and Nancy Kaehr. This research was supported in part by the
California Institute of Regenerative Medicine (RS1-00172-1) to S.C. and (RB1-01406) to K.W.
Cellular and Molecular Medicine, Stem Cell Program, University of California, San Diego, California, USA
Department of Bioengineering, University of California, San Diego, California, USA
D.A.B., S.C. and K.W. developed the protocol. D.A.B. and K.W. designed and performed the experiments. D.A.B., S.C. and K.W. analyzed the results. D.A.B., S.C. and K.W. wrote the manuscript.
Schematic representation of ACME format. (a) Schematic of a 10x10 subarray that contains 20 different conditions spotted in replicates of five. Spot diameter (closed circles) is 150 μm and
the center to center distance between neighboring spots is 450 μm. These subarrays can be arranged to generate several different designs including (b) a 8x2, 1600 spot/320 condition, (c) a
16x4, 6400 spot/1280 condition, or (d) a 16x5, 8000 spot/1600 condition array. (PPT 415 kb)
Example of analysis of raw data generated from the array in Figure 5e. (DOC 1214 kb)
Anyone you share the following link with will be able to read this content: