Genetic architecture and evolution of the S locus supergene in Primula vulgaris
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
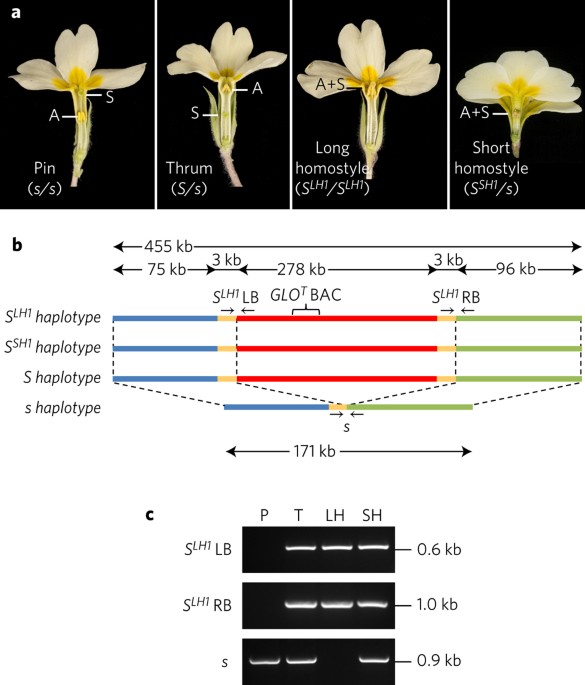
Darwin's studies on heterostyly in Primula described two floral morphs, pin and thrum, with reciprocal anther and stigma heights that promote insect-mediated cross-pollination. This key
innovation evolved independently in several angiosperm families. Subsequent studies on heterostyly in Primula contributed to the foundation of modern genetic theory and the neo-Darwinian
synthesis. The established genetic model for Primula heterostyly involves a diallelic S locus comprising several genes, with rare recombination events that result in self-fertile homostyle
flowers with anthers and stigma at the same height. Here we reveal the S locus supergene as a tightly linked cluster of thrum-specific genes that are absent in pins. We show that thrums are
hemizygous not heterozygous for the S locus, which suggests that homostyles do not arise by recombination between S locus haplotypes as previously proposed. Duplication of a floral homeotic
gene 51.7 million years (Myr) ago, followed by its neofunctionalization, created the current S locus assemblage which led to floral heteromorphy in Primula. Our findings provide new insights
into the structure, function and evolution of this archetypal supergene.
We thank M. Lappage, M. Hughes and P. Wells for horticultural support; colleagues at TGAC for Illumina sequencing; A. Thanki for TGAC Browser support; O. Kent for P. elatior GLO and GLOT
sequences; Norfolk Wildlife Trust, Suffolk Wildlife Trust and Norfolk County Council for permission to sample P. veris, P. elatior and P. vulgaris respectively; M. Gage, B. Davies and D.
Bowles for comments on the manuscript; W. Wang for advice on k-means analysis; BBSRC for funding via grant BB/H019278/2, and prior awards G11027 and P11021; The Gatsby Foundation for early
stage funding; University of Leeds, Durham University and University of East Anglia for support to P.M.G. over several years of the project. CvO was funded by the Earth & Life Systems
Alliance (ELSA). P.M.G.'s laboratory is hosted at the John Innes Centre under the UEA-JIC Norwich Research Park collaboration.
Present address: †Present address: National Institute for Agricultural Botany, Huntingdon Road, Cambridge CB3 0LE, UK,
Jinhong Li and Jonathan M. Cocker: These authors contributed equally to this work.
School of Biological Sciences, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK
Jinhong Li, Jonathan M. Cocker, Margaret A. Webster & Philip M. Gilmartin
John Innes Centre, Norwich Research Park, Norwich, NR4 7UH, UK
Jinhong Li, Jonathan M. Cocker, Margaret A. Webster & Philip M. Gilmartin
The Earlham Institute, Norwich Research Park, Norwich, NR4 7UH, UK
Jonathan Wright, Mark McMullan, Sarah Dyer, David Swarbreck & Mario Caccamo
School of Environmental Sciences, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK
J.L. contributed to project design, performed all molecular analyses, generated the S locus assembly, manually annotated the S locus gene structures and undertook data analysis. J.M.C.
carried out bioinformatic analyses, including automated annotation of the S locus region, undertook in silico gene expression and k-means clustering analyses, assembled genome sequences and
library scaffolds, generated the molecular phylogeny, undertook recombination analysis of the S locus flanking regions and contributed to project design. J.W. assembled genome sequences and
library scaffolds, contributed to genome annotation and generated the automated gene model predictions across the S locus, aligned sequencing reads to the S locus assembly and contributed to
project design. M.A.W. contributed the inbred long homostyle line, other genetic resources and classical genetics, identified the short homostyle mutant and generated the three-point cross
used to demonstrate linkage. M.M. and C.v.O. contributed to the molecular phylogeny construction, evolutionary data analysis and recombination analysis. S.A., D.S. and M.C. contributed to
the genome sequencing strategy, assembly and annotation that underpins this project. P.M.G. conceived, designed and directed the project, contributed to data analysis, prepared the figures
and drafted the manuscript, with revision input from C.v.O.; all authors contributed to editing the manuscript.
Supplementary Methods, Supplementary References, Supplementary Figures 1–4, Supplementary Tables 1–6, Supplementary Sequence Analyses 1–3. (PDF 1701 kb)
Anyone you share the following link with will be able to read this content: