Intensity normalization improves color calling in solid sequencing
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Access through your institution Buy or subscribe To the Editor: Applied Biosystems' SOLiD system1 is a commonly used massively parallel DNA sequencing platform for applications from
genotyping and structural variation analysis1 to transcriptome quantification and reconstruction2. Like other sequencing technologies, it measures fluorescence intensities from dye-labeled
molecules to determine the sequence of DNA fragments. Ultimately, sequences are determined by complicated statistical manipulations of noisy intensity measurements, and systematic biases may
mislead downstream analysis3. Several proposed methods improve base calling and quality metrics for other sequencing technologies3,4,5, and we now present Rsolid, software implementing an
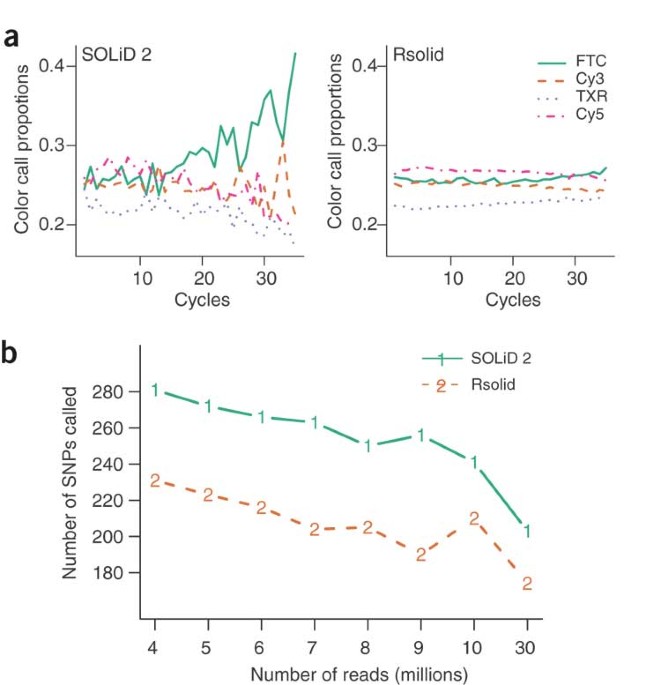
intensity normalization strategy for the SOLiD platform that substantially improves yield and accuracy at small computational costs (6% increase in total matches, 13% increase in perfect
matches, 5% reduced error rate and a substantial reduction in false positive single-nucleotide polymorphism (SNP) calls in an _Escherichia coli_ genomic DNA sample). This is a preview of
subscription content, access via your institution RELEVANT ARTICLES Open Access articles citing this article. * PERFORMANCE ASSESSMENT OF UP-FLOW ANAEROBIC MULTI-STAGED REACTOR FOLLOWED BY
AUTO-AERATED IMMOBILIZED BIOMASS UNIT FOR TREATING POLYESTER WASTEWATER, WITH BIOGAS PRODUCTION * Raouf Hassan * , Karim Kriaa * … Ahmed Tawfik _Applied Water Science_ Open Access 14 March
2024 * COMPARATIVE TRANSCRIPTOME ANALYSIS OF CHINARY, ASSAMICA AND CAMBOD TEA (CAMELLIA SINENSIS) TYPES DURING DEVELOPMENT AND SEASONAL VARIATION USING RNA-SEQ TECHNOLOGY * Ajay Kumar * ,
Vandna Chawla * … Sudesh Kumar Yadav _Scientific Reports_ Open Access 17 November 2016 * COMPREHENSIVE TRANSCRIPTOMIC STUDY ON HORSE GRAM (MACROTYLOMA UNIFLORUM): DE NOVO ASSEMBLY,
FUNCTIONAL CHARACTERIZATION AND COMPARATIVE ANALYSIS IN RELATION TO DROUGHT STRESS * Jyoti Bhardwaj * , Rohit Chauhan * … Sudesh Kumar Yadav _BMC Genomics_ Open Access 23 September 2013
ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article *
Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn
about institutional subscriptions * Read our FAQs * Contact customer support REFERENCES * McKernan, K.J., et al. _Genome Res._ 19, 1527–1541 (2009). Article CAS Google Scholar * Tang, F.
et al. _Nat. Methods_ 6, 377–382 (2009). Article CAS Google Scholar * Bravo, H.C. & Irizarry, R.A. _Biometrics_ advance online publication, 13 November 2009 (doi:
10.1111/j.1541-0420.2009.01353.x). Article Google Scholar * Quinlan, A.R., Stewart, D.A., Stromberg, M.P. & Marth, G. _Nat. Methods_ 5, 179–181 (2008). Article CAS Google Scholar *
Kao, W.C., Stevens, K. & Song, Y.S. _Genome Res._ 19, 1884–1895 (2009). Article CAS Google Scholar * Hayden, E.C. _Nature_ 451, 378–379 (2008). Article Google Scholar Download
references AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA Hao Wu, Rafael A Irizarry
& Héctor Corrada Bravo Authors * Hao Wu View author publications You can also search for this author inPubMed Google Scholar * Rafael A Irizarry View author publications You can also
search for this author inPubMed Google Scholar * Héctor Corrada Bravo View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS
Correspondence to Hao Wu or Héctor Corrada Bravo. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT
AND FIGURES Supplementary Figure 1, Supplementary Tables 1–4, Supplementary Methods (PDF 1021 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Wu, H.,
Irizarry, R. & Bravo, H. Intensity normalization improves color calling in SOLiD sequencing. _Nat Methods_ 7, 336–337 (2010). https://doi.org/10.1038/nmeth0510-336 Download citation *
Issue Date: May 2010 * DOI: https://doi.org/10.1038/nmeth0510-336 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative