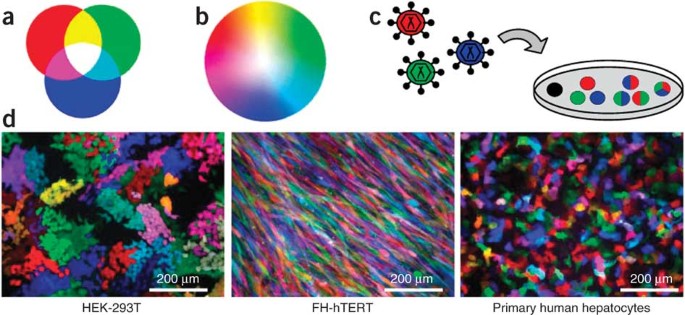
Rgb marking facilitates multicolor clonal cell tracking
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT We simultaneously transduced cells with three lentiviral gene ontology (LeGO) vectors encoding red, green or blue fluorescent proteins. Individual cells were thereby marked by
different combinations of inserted vectors, resulting in the generation of numerous mixed colors, a principle we named red-green-blue (RGB) marking. We show that lentiviral vector–mediated
RGB marking remained stable after cell division, thus facilitating the analysis of clonal cell fates _in vitro_ and _in vivo_. Particularly, we provide evidence that RGB marking allows
assessment of clonality after regeneration of injured livers by transplanted primary hepatocytes. We also used RGB vectors to mark hematopoietic stem/progenitor cells that generated colored
spleen colonies. Finally, based on limiting-dilution and serial transplantation assays with tumor cells, we found that clonal tumor cells retained their specific color-code over extensive
periods of time. We conclude that RGB marking represents a useful tool for cell clonality studies in tissue regeneration and pathology. Access through your institution Buy or subscribe This
is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00
per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated
during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS INTERROGATION
OF CLONAL TRACKING DATA USING BARCODETRACKR Article 26 April 2021 IFLPMOSAICS ENABLE THE MULTISPECTRAL BARCODING AND HIGH-THROUGHPUT COMPARATIVE ANALYSIS OF MUTANT AND WILD-TYPE CELLS
Article Open access 13 December 2024 ISOLATING LIVE CELL CLONES FROM BARCODED POPULATIONS USING CRISPRA-INDUCIBLE REPORTERS Article 27 July 2020 REFERENCES * Shaner, N.C., Steinbach, P.
& Tsien, R. A guide to choosing fluorescent proteins. _Nat. Methods_ 2, 905–909 (2005). Article CAS Google Scholar * Giepmans, B.N., Adams, S.R., Ellisman, M.H. & Tsien, R.Y. The
fluorescent toolbox for assessing protein location and function. _Science_ 312, 217–224 (2006). Article CAS Google Scholar * Vafaizadeh, V. et al. Mammary epithelial reconstitution with
gene-modified stem cells assigns roles to Stat5 in luminal alveolar cell fate decisions, differentiation, involution and mammary tumor formation. _Stem Cells_ 28, 928–938 (2010). CAS PubMed
Google Scholar * Yamauchi, K. et al. Development of real-time subcellular dynamic multicolor imaging of cancer-cell trafficking in live mice with a variable-magnification whole-mouse
imaging system. _Cancer Res._ 66, 4208–4214 (2006). Article CAS Google Scholar * Tysnes, B.B. Tumor-initiating and -propagating cells: cells that we would like to identify and control.
_Neoplasia_ 12, 506–515 (2010). Article CAS Google Scholar * Dick, J.E. Stem cell concepts renew cancer research. _Blood_ 112, 4793–4807 (2008). Article CAS Google Scholar * Livet, J.
et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. _Nature_ 450, 56–62 (2007). Article CAS Google Scholar * Weber, K., Bartsch, U.,
Stocking, C. & Fehse, B. A multi-color panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. _Mol. Ther._ 16, 698–706 (2008). Article CAS Google
Scholar * Shaner, N.C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from _Discosoma_ sp. red fluorescent protein. _Nat. Biotechnol._ 22, 1567–1572 (2004).
Article CAS Google Scholar * Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. _Nat. Biotechnol._ 20, 87–90
(2002). Article CAS Google Scholar * Rizzo, M.A., Springer, G., Granada, B. & Piston, D. An improved cyan fluorescent protein variant useful for FRET. _Nat. Biotechnol._ 22, 445–449
(2004). Article CAS Google Scholar * Kustikova, O.S. et al. Dose finding with retroviral vectors: Correlation of retroviral vector copy numbers in single cells with gene transfer
efficiency in a cell population. _Blood_ 102, 3934–3937 (2003). Article CAS Google Scholar * Fehse, B., Kustikova, O.S., Bubenheim, M. & Baum, C. Pois(s)on—it's a question of
dose.... _Gene Ther._ 11, 879–881 (2004). Article CAS Google Scholar * Wege, H. et al. Telomerase reconstitution immortalizes human fetal hepatocytes without disrupting their
differentiation potential. _Gastroenterology_ 124, 432–444 (2003). Article CAS Google Scholar * Dandri, M. et al. Repopulation of mouse liver with human hepatocytes and in vivo infection
with hepatitis B virus. _Hepatology_ 33, 981–988 (2001). Article CAS Google Scholar * Petersen, J. et al. Prevention of hepatitis B virus infection _in vivo_ by entry inhibitors derived
from the large envelope protein. _Nat. Biotechnol._ 26, 335–341 (2008). Article CAS Google Scholar * Hock, H. Some hematopoietic stem cells are more equal than others. _J. Exp. Med._ 207,
1127–1130 (2010). Article CAS Google Scholar * Metcalf, D. et al. Two distinct types of murine blast colony-forming cells are multipotential hematopoietic precursors. _Proc. Natl. Acad.
Sci. USA_ 105, 18501–18506 (2008). Article CAS Google Scholar * Townsend, C.M. Jr., Ishizuka, J. & Thompson, J.C. Studies of growth regulation in a neuroendocrine cell line. _Acta
Oncol._ 32, 125–130 (1993). Article Google Scholar * Naldini, L. et al. _In vivo_ gene delivery and stable transduction of nondividing cells by a lentiviral vector. _Science_ 272, 263–267
(1996). Article CAS Google Scholar * Rieger, M.A., Hoppe, P.S., Smejkal, B.M., Eitelhuber, A.C. & Schroeder, T. Hematopoietic cytokines can instruct lineage choice. _Science_ 325,
217–218 (2009). Article CAS Google Scholar * Cui, K. et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation.
_Cell Stem Cell_ 4, 80–93 (2009). Article CAS Google Scholar * Stein, S. et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for
chronic granulomatous disease. _Nat. Med._ 16, 198–204 (2010). Article CAS Google Scholar * Kim, Y.J. et al. Sustained high-level polyclonal hematopoietic marking and transgene
expression 4 years after autologous transplantation of rhesus macaques with SIV lentiviral vector–transduced CD34+ cells. _Blood_ 113, 5434–5443 (2009). Article CAS Google Scholar *
Aiuti, A. et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. _N. Engl. J. Med._ 360, 447–458 (2009). Article CAS Google Scholar * Lütgehetmann, M. et al. _In
vivo_ proliferation of hepadnavirus-infected hepatocytes induces loss of covalently closed circular DNA in mice. _Hepatology_ 52, 16–24 (2010). Article Google Scholar * Novelli, M. et al.
X-inactivation patch size in human female tissue confounds the assessment of tumor clonality. _Proc. Natl. Acad. Sci. USA_ 100, 3311–3314 (2003). Article CAS Google Scholar * Leedham,
S.J. & Wright, N.A. Human tumour clonality assessment—flawed but necessary. _J. Pathol._ 215, 351–354 (2008). Article CAS Google Scholar * Weber, K., Mock, U., Petrowitz, B., Bartsch,
U. & Fehse, B. LeGO vectors equipped with novel drug-selectable fluorescent proteins—new building blocks for cell marking and multi-gene analysis. _Gene Ther._ 17, 511–520 (2010).
Article CAS Google Scholar * Beyer, W.R., Westphal, M., Ostertag, W. & von Laer, D. Oncoretrovirus and lentivirus vectors pseudotyped with lymphocytic choriomeningitis virus
glycoprotein: generation, concentration and broad host range. _J. Virol._ 76, 1488–1495 (2002). Article CAS Google Scholar * Li, W.C., Ralphs, K.L. & Tosh, D. Isolation and culture of
adult mouse hepatocytes. _Methods Mol. Biol._ 633, 185–196 (2010). Article CAS Google Scholar * Benten, D. et al. Hepatic targeting of transplanted liver sinusoidal endothelial cells in
intact mice. _Hepatology_ 42, 140–148 (2005). Article CAS Google Scholar * Schüler, A. et al. The MADS transcription factor Mef2c is a pivotal modulator of myeloid cell fate. _Blood_ 111,
4532–4541 (2008). Article Google Scholar * Schwieger, M. et al. Homing and invasiveness of MLL/ENL leukemic cells is regulated by MEF2C. _Blood_ 114, 2476–2488 (2009). Article CAS
Google Scholar * Weber, K & Fehse, B. Diva-Fit: a step-by-step manual for generating high-resolution graphs and histogram overlays of flow cytometry data obtained with FACSDiva
software. _Cell. Ther. Transplant._ 1, e.000045.01 (2009). Google Scholar * Kustikova, O.S., Baum, C. & Fehse, B. Retroviral integration site analysis in hematopoietic stem cells.
_Methods Mol. Biol._ 430, 255–267 (2008). Article CAS Google Scholar * Kustikova, O.S., Modlich, U. & Fehse, B. Retroviral insertion site analysis in dominant haematopoietic clones.
_Methods Mol. Biol._ 506, 373–390 (2009). Article CAS Google Scholar * Cornils, K. et al. Stem cell marking with promotor-deprived self-inactivating retroviral vectors does not lead to
induced clonal imbalance. _Mol. Ther._ 17, 131–143 (2009). Article CAS Google Scholar * Appelt, J.U. et al. QuickMap: a public tool for large-scale gene therapy vector insertion site
mapping and analysis. _Gene Ther._ 16, 885–893 (2009). Article CAS Google Scholar * Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers.
_Methods Mol. Biol._ 132, 365–386 (2000). CAS Google Scholar Download references ACKNOWLEDGEMENTS We wish to thank to V. Matzat and U. Bergholz for expert technical assistance, R. Reusch
for excellent mouse care and J. Petersen for continuous support with the uPA model and critical reading of the manuscript. Confocal imaging was performed with the kind help from O. Bruns in
collaboration with the Nikon Application Center Northern Germany (Nikon). We are indebted to many colleagues for their kind support with various cells and constructs: H. Wege (University
Medical Center Hamburg-Eppendorf) for FH-hTERT, D. Hösch (University Marburg) for BON cells, W. Beyer (Heinrich-Pette-Institute) for vesicular stomatitis virus G protein cDNA R.Y. Tsien
(Howard Hughes Medical Institute) for mCherry cDNA, A. Miyawaki (RIKEN) and T. Schroeder (Institute for Stem Cell Research) for Venus cDNA and D.W. Piston (Vanderbilt-Ingram Cancer Center)
for Cerulean cDNA. This work was supported by the Deutsche Forschungsgemeinschaft (SFB841 to B.F., D.B. and M.D. and FE568/11-1 to B.F.). M. Thomaschewski was supported by the integrated
graduate school “Inflammation and Regeneration” of the University Medical Center Hamburg-Eppendorf. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Research Department Cell and Gene Therapy,
Clinic for Stem Cell Transplantation, University Cancer Center Hamburg, University Medical Center Hamburg-Eppendorf, Hamburg, Germany Kristoffer Weber, Michael Thomaschewski, Michael
Warlich, Kerstin Cornils & Boris Fehse * Internal Medicine I, University Medical Center Hamburg-Eppendorf, Hamburg, Germany Michael Warlich, Tassilo Volz, Marc Lütgehetmann, Maura Dandri
& Daniel Benten * Heinrich-Pette-Institute, Leibniz-Institute for Experimental Virology, Hamburg, Germany Birte Niebuhr, Maike Täger & Carol Stocking * Department of Hepatobiliary
and Transplant Surgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany Jörg-Matthias Pollok Authors * Kristoffer Weber View author publications You can also search for this
author inPubMed Google Scholar * Michael Thomaschewski View author publications You can also search for this author inPubMed Google Scholar * Michael Warlich View author publications You can
also search for this author inPubMed Google Scholar * Tassilo Volz View author publications You can also search for this author inPubMed Google Scholar * Kerstin Cornils View author
publications You can also search for this author inPubMed Google Scholar * Birte Niebuhr View author publications You can also search for this author inPubMed Google Scholar * Maike Täger
View author publications You can also search for this author inPubMed Google Scholar * Marc Lütgehetmann View author publications You can also search for this author inPubMed Google Scholar
* Jörg-Matthias Pollok View author publications You can also search for this author inPubMed Google Scholar * Carol Stocking View author publications You can also search for this author
inPubMed Google Scholar * Maura Dandri View author publications You can also search for this author inPubMed Google Scholar * Daniel Benten View author publications You can also search for
this author inPubMed Google Scholar * Boris Fehse View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS K.W. designed the study, produced LeGO
vectors and performed and analyzed gene transfer experiments in target cells _in vitro_ and _in vivo_. M. Thomaschewski and M.W. isolated, transduced and transplanted primary hepatocytes and
analyzed mice. M. Thomaschewski also performed mouse studies with BON tumor cells. T.V. and M.L. performed experiments in uPA mice. K.C. identified vector insertions by LM-PCR and performed
specific PCRs. B.N., M. Täger and C.S. designed and performed experiments with mouse HSCs. J.-M.P. and M.L. provided and prepared primary human hepatocytes. M.D. provided the uPA-SCID model
and supervised _in vivo_ experiments in that model. D.B. designed and performed mouse studies. B.F. designed the study, analyzed and evaluated results and wrote the manuscript. All authors
read and approved the final version of the manuscript. CORRESPONDING AUTHOR Correspondence to Boris Fehse. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial
interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Figures 1–4 and Supplementary Tables 1 and 2 (PDF 662 kb) RIGHTS AND PERMISSIONS Reprints and permissions
ABOUT THIS ARTICLE CITE THIS ARTICLE Weber, K., Thomaschewski, M., Warlich, M. _et al._ RGB marking facilitates multicolor clonal cell tracking. _Nat Med_ 17, 504–509 (2011).
https://doi.org/10.1038/nm.2338 Download citation * Received: 10 May 2010 * Accepted: 01 December 2010 * Published: 27 March 2011 * Issue Date: April 2011 * DOI:
https://doi.org/10.1038/nm.2338 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative