The study of macromolecular complexes by quantitative proteomics
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
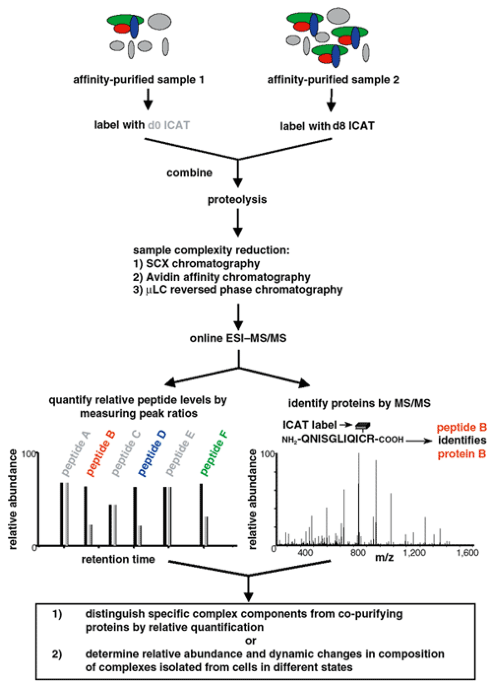
ABSTRACT We describe a generic strategy for determining the specific composition, changes in the composition, and changes in the abundance of protein complexes. It is based on the use of
isotope-coded affinity tag (ICAT) reagents1 and mass spectrometry to compare the relative abundances of tryptic peptides derived from suitable pairs of purified or partially purified protein
complexes. In a first application, the genuine protein components of a large RNA polymerase II (Pol II) preinitiation complex (PIC) were distinguished from a background of co-purifying
proteins by comparing the relative abundances of peptides derived from a control sample and the specific complex that was purified from nuclear extracts by a single-step promoter DNA
affinity procedure2. In a second application, peptides derived from immunopurified STE12 protein complexes isolated from yeast cells in different states were used to detect quantitative
changes in the abundance of the complexes, and to detect dynamic changes in the composition of the samples. The use of quantitative mass spectrometry to guide identification of specific
complex components in partially purified samples, and to detect quantitative changes in the abundance and composition of protein complexes, provides the researcher with powerful new tools
for the comprehensive analysis of macromolecular complexes. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS
OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on
SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about
institutional subscriptions * Read our FAQs * Contact customer support REFERENCES * Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags.
_Nat. Biotechnol._ 17, 994–999 (1999). Article CAS Google Scholar * Ranish, J.A., Yudkovsky, N. & Hahn, S. Intermediates in formation and activity of the RNA polymerase II
preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. _Genes Dev._ 13, 49–63 (1999). Article CAS Google Scholar * Hartwell, L.H., Hopfield,
J.J., Leibler, S. & Murray, A.W. From molecular to modular cell biology. _Nature_ 402, C47–C52 (1999). Article CAS Google Scholar * Fields, S. & Song, O.-k. A novel genetic system
to detect protein–protein interactions. _Nature_ 340, 245–246 (1989). Article CAS Google Scholar * Uetz, P. et al. A comprehensive analysis of protein–protein interactions in
_Saccharomyces cerevisiae_. _Nature_ 403, 623–627 (2000). Article CAS Google Scholar * Gavin, A.C. et al. Functional organization of the yeast proteome by systematic analysis of protein
complexes. _Nature_ 415, 141–147 (2002). Article CAS Google Scholar * Ho, Y. et al. Systematic identification of protein complexes in _Saccharomyces cerevisiae_ by mass spectrometry.
_Nature_ 415, 180–183 (2002). Article CAS Google Scholar * Zhou, H., Ranish, J.A., Watts, J.D. & Aebersold, R. Quantitative proteome analysis by solid-phase isotope tagging and mass
spectrometry. _Nat. Biotechnol._ 20, 512–515 (2002). Article CAS Google Scholar * Cagney, G. & Emili, A. _De novo_ peptide sequencing and quantitative profiling of complex protein
mixtures using mass-coded abundance tagging. _Nat. Biotechnol._ 20, 163–170 (2002). Article CAS Google Scholar * Han, D.K., Eng, J., Zhou, H. & Aebersold, R. Quantitative profiling of
differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. _Nat. Biotechnol._ 19, 946–951 (2001). Article CAS Google Scholar * Eng, J.K.,
McCormack, A.L. & Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in protein databases. _J. Am. Soc. Mass Spectrom._ 5, 976–989
(1994). Article CAS Google Scholar * Lee, T.I. & Young, R.A. Transcription of eukaryotic protein-coding genes. _Annu. Rev. Genet._ 34, 77–137 (2000). Article CAS Google Scholar *
Li, X.Y., Bhaumik, S.R. & Green, M.R. Distinct classes of yeast promoters revealed by differential TAF recruitment. _Science_ 288, 1242–1244 (2000). Article CAS Google Scholar *
Kuras, L., Kosa, P., Mencia, M. & Struhl, K. TAF-containing and TAF-independent forms of transcriptionally active TBP _in vivo_. _Science_ 288, 1244–1248 (2000). Article CAS Google
Scholar * Fry, C.J. & Peterson, C.L. Chromatin remodeling enzymes: who's on first? _Curr. Biol._ 11, R185–R197 (2001). Article CAS Google Scholar * Reddy, P. & Hahn, S.
Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. _Cell_ 65, 349–357 (1991). Article CAS Google Scholar * Costanzo, M.C. et al. YPD, PombePD and WormPD:
model organism volumes of the BioKnowledge library, an integrated resource for protein information. _Nucleic Acids Res._ 29, 75–79 (2001). Article CAS Google Scholar * Zhang, X., Jin,
Q.K., Carr, S.A. & Annan, R.S. N-terminal peptide labeling strategy for incorporation of isotopic tags: a method for the determination of site-specific absolute phosphorylation
stoichiometry. _Rapid Commun. Mass Spectrom._ 16, 2325–2332 (2002). Article CAS Google Scholar * Yudkovsky, N., Ranish, J.A. & Hahn, S. A transcription reinitiation intermediate that
is stabilized by activator. _Nature_ 408, 225–229 (2000). Article CAS Google Scholar * Auble, D.T. et al. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding
to DNA by an ATP-dependent mechanism. _Genes Dev._ 8, 1920–1934 (1994). Article CAS Google Scholar * Sprague, G.F. & Thorner, J.W. Pherormone response and signal transduction during
the mating process of _Saccharomyces cerevisiae_. in _The Molecular and Cellular Biology of the Yeast Saccharomyces_ Vol. 2 (eds. Jones, E.W., Pringle, J.R. & Broach, J.R.) 657–744 (Cold
Spring Harbor Laboratory Press, Plainview, 1992). Google Scholar * Tedford, K., Kim, S., Sa, D., Stevens, K. & Tyers, M. Regulation of the mating pheromone and invasive growth
responses in yeast by two MAP kinase substrates. _Curr. Biol._ 7, 228–238 (1997). Article CAS Google Scholar * Olson, M.V. Genome structure and organization in _Saccharomyces cerevisiae_.
in _The Molecular and Cellular Biology of the Yeast Saccharomyces_ Vol. 1 (eds. Broach, J.R., Pringle, J. & Jones, E.) 1–39 (Cold Spring Harbor Laboratory Press, Plainview, 1991).
Google Scholar * Roberts, C.J. et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. _Science_ 287, 873–880 (2000). Article CAS
Google Scholar * Griffin, T.J. et al. Quantitative proteomic analysis using a MALDI quadrupole time-of-flight mass spectrometer. _Anal. Chem._ 73, 978–986 (2001). Article CAS Google
Scholar * Griffin, T.J. et al. Toward a high-throughput approach to quantitative proteomic analysis: expression-dependent protein identification by mass spectrometry. _J. Am. Soc. Mass
Spectrom._ 12, 1238–1246 (2001). Article CAS Google Scholar * Shiio, Y. et al. Quantitative proteomic analysis of Myc oncoprotein function. _EMBO J._ 21, 5088–5096 (2002). Article CAS
Google Scholar * Kang, J.J., Auble, D.T., Ranish, J.A. & Hahn, S. Analysis of the yeast transcription factor TFIIA: distinct functional regions and a polymerase II-specific role in
basal and activated transcription. _Mol. Cell. Biol._ 15, 1234–1243 (1995). Article CAS Google Scholar * Aitchison, J.D., Rout, M.P., Marelli, M., Blobel, G. & Wozniak, R.W. Two novel
related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. _J.
Cell Biol._ 131, 1133–1148 (1995). Article CAS Google Scholar * Yi, E.C. et al. Approaching complete peroxisome characterization by gas-phase fractionation. _Electrophoresis_ 23,
3205–3216 (2002). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank S. Hahn for the gift of rTBP and antibodies to TBP, TFIIB and SRB4, A. Nesvizhskii for help with
data analysis and J. Aitchison, M. Wright and B. Wollscheid for comments on the manuscript. This work was supported by grants from the US National Cancer Institute and US National
Institutes of Health Research Resource Center, by federal funds from the National Heart, Lung, and Blood Institute of the National Institutes of Health and by a postdoctoral fellowship from
the National Institutes of Health to J.A.R. Partial funding for this work came through a gift from Merck and Co. to the Institute for Systems Biology. AUTHOR INFORMATION Author notes *
Samuel O. Purvine Present address: Biatech, 19310 North Creek Parkway, Suite 115, Bothell, Washington, 98011, USA AUTHORS AND AFFILIATIONS * Institute for Systems Biology, 1441 North 34th
Street, Seattle, 98103-8904, Washington, USA Jeffrey A. Ranish, Eugene C. Yi, Deena M. Leslie, David R. Goodlett, Jimmy Eng & Ruedi Aebersold Authors * Jeffrey A. Ranish View author
publications You can also search for this author inPubMed Google Scholar * Eugene C. Yi View author publications You can also search for this author inPubMed Google Scholar * Deena M. Leslie
View author publications You can also search for this author inPubMed Google Scholar * Samuel O. Purvine View author publications You can also search for this author inPubMed Google Scholar
* David R. Goodlett View author publications You can also search for this author inPubMed Google Scholar * Jimmy Eng View author publications You can also search for this author inPubMed
Google Scholar * Ruedi Aebersold View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Ruedi Aebersold. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIG. 1 SUPPLEMENTARY TABLE 1 SUPPLEMENTARY TABLE 2 RIGHTS AND
PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Ranish, J., Yi, E., Leslie, D. _et al._ The study of macromolecular complexes by quantitative proteomics. _Nat
Genet_ 33, 349–355 (2003). https://doi.org/10.1038/ng1101 Download citation * Received: 07 October 2002 * Accepted: 19 January 2003 * Published: 18 February 2003 * Issue Date: March 2003 *
DOI: https://doi.org/10.1038/ng1101 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative