Exome sequencing supports a de novo mutational paradigm for schizophrenia
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Despite its high heritability, a large fraction of individuals with schizophrenia do not have a family history of the disease (sporadic cases). Here we examined the possibility that
rare _de novo_ protein-altering mutations contribute to the genetic component of schizophrenia by sequencing the exomes of 53 sporadic cases, 22 unaffected controls and their parents. We
identified 40 _de novo_ mutations in 27 cases affecting 40 genes, including a potentially disruptive mutation in _DGCR2_, a gene located in the schizophrenia-predisposing 22q11.2
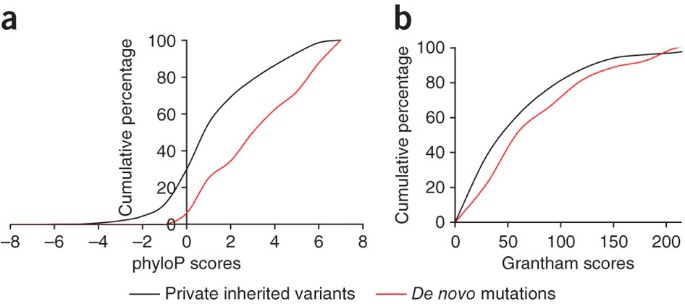
microdeletion region. A comparison to rare inherited variants indicated that the identified _de novo_ mutations show a large excess of non-synonymous changes in schizophrenia cases, as well
as a greater potential to affect protein structure and function. Our analyses suggest a major role for _de novo_ mutations in schizophrenia as well as a large mutational target, which
together provide a plausible explanation for the high global incidence and persistence of the disease. Access through your institution Buy or subscribe This is a preview of subscription
content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue
Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL
ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS THE MOLECULAR PATHOLOGY OF SCHIZOPHRENIA:
AN OVERVIEW OF EXISTING KNOWLEDGE AND NEW DIRECTIONS FOR FUTURE RESEARCH Article Open access 06 March 2023 SCHIZOPHRENIA RISK CONFERRED BY RARE PROTEIN-TRUNCATING VARIANTS IS CONSERVED
ACROSS DIVERSE HUMAN POPULATIONS Article Open access 13 March 2023 THE GENETIC ARCHITECTURE OF SCHIZOPHRENIA: REVIEW OF LARGE-SCALE GENETIC STUDIES Article 12 July 2022 ACCESSION CODES
ACCESSIONS GENBANK/EMBL/DDBJ * NM_000090 * NM_000110 * NM_000426 * NM_001004703 * NM_001037162 * NM_001042646 * NM_001083591 * NM_001114 * NM_001130848 * NM_001144382 * NM_001145025 *
NM_002675 * NM_002926 * NM_003246 * NM_003496 * NM_004958 * NM_005137 * NM_005142 * NM_005462 * NM_005539 * NM_006545 * NM_007249 * NM_013260 * NM_014243 * NM_014781 * NM_018117 * NM_018206
* NM_018440 * NM_019093 * NM_020880 * NM_021826 * NM_052961 * NM_134266 * NM_138805 * NM_138961 * NM_144674 * NM_145207 * NM_152389 * NM_153838 * NM_207370 REFERENCES * Gottesman, I.I. &
Shields, J. A polygenic theory of schizophrenia. _Proc. Natl. Acad. Sci. USA_ 58, 199–205 (1967). Article CAS Google Scholar * Sullivan, P.F., Kendler, K.S. & Neale, M.C.
Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. _Arch. Gen. Psychiatry_ 60, 1187–1192 (2003). Article Google Scholar * Lichtenstein, P. et al. Common
genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. _Lancet_ 373, 234–239 (2009). Article CAS Google Scholar * Lupski, J.R. Genomic
rearrangements and sporadic disease. _Nat. Genet._ 39, S43–S47 (2007). Article CAS Google Scholar * Karayiorgou, M. et al. Schizophrenia susceptibility associated with interstitial
deletions of chromosome 22q11. _Proc. Natl. Acad. Sci. USA_ 92, 7612–7616 (1995). Article CAS Google Scholar * Xu, B. et al. Strong association of _de novo_ copy number mutations with
sporadic schizophrenia. _Nat. Genet._ 40, 880–885 (2008). Article CAS Google Scholar * Cirulli, E.T. & Goldstein, D.B. Uncovering the roles of rare variants in common disease through
whole-genome sequencing. _Nat. Rev. Genet._ 11, 415–425 (2010). Article CAS Google Scholar * O'Roak, B.J. et al. Exome sequencing in sporadic autism spectrum disorders identifies
severe _de novo_ mutations. _Nat. Genet._ 43, 585–589 (2011). Article CAS Google Scholar * Vissers, L.E. et al. A _de novo_ paradigm for mental retardation. _Nat. Genet._ 42, 1109–1112
(2010). Article CAS Google Scholar * Awadalla, P. et al. Direct measure of the _de novo_ mutation rate in autism and schizophrenia cohorts. _Am. J. Hum. Genet._ 87, 316–324 (2010).
Article CAS Google Scholar * Abecasis, G.R. et al. Genomewide scan in families with schizophrenia from the founder population of Afrikaners reveals evidence for linkage and uniparental
disomy on chromosome 1. _Am. J. Hum. Genet._ 74, 403–417 (2004). Article CAS Google Scholar * Karayiorgou, M. et al. Phenotypic characterization and genealogical tracing in an Afrikaner
schizophrenia database. _Am. J. Med. Genet. B Neuropsychiatr. Genet._ 124B, 20–28 (2004). Article Google Scholar * Xu, B. et al. Elucidating the genetic architecture of familial
schizophrenia using rare copy number variant and linkage scans. _Proc. Natl. Acad. Sci. USA_ 106, 16746–16751 (2009). Article Google Scholar * Li, H. & Durbin, R. Fast and accurate
short read alignment with Burrows-Wheeler transform. _Bioinformatics_ 25, 1754–1760 (2009). Article CAS Google Scholar * Bentley, D.R. et al. Accurate whole human genome sequencing using
reversible terminator chemistry. _Nature_ 456, 53–59 (2008). Article CAS Google Scholar * 1000 Genomes Project Consortium. et al. A map of human genome variation from population-scale
sequencing. _Nature_ 467, 1061–1073 (2010). * Lynch, M. Rate, molecular spectrum, and consequences of human mutation. _Proc. Natl. Acad. Sci. USA_ 107, 961–968 (2010). Article CAS Google
Scholar * Li, Y. et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. _Nat. Genet._ 42, 969–972 (2010). Article CAS Google Scholar
* Botstein, D. & Risch, N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. _Nat. Genet._ 33 (suppl.),
228–237 (2003). Article CAS Google Scholar * Pollard, K.S., Hubisz, M.J., Rosenbloom, K.R. & Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. _Genome
Res._ 20, 110–121 (2010). Article CAS Google Scholar * Grantham, R. Amino acid difference formula to help explain protein evolution. _Science_ 185, 862–864 (1974). Article CAS Google
Scholar * Kajiwara, K. et al. Cloning of _SEZ-12_ encoding seizure-related and membrane-bound adhesion protein. _Biochem. Biophys. Res. Commun._ 222, 144–148 (1996). Article CAS Google
Scholar * Karayiorgou, M., Simon, T.J. & Gogos, J.A. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. _Nat. Rev. Neurosci._ 11, 402–416
(2010). Article CAS Google Scholar * Bjarnadótttir, T.K. et al. The human and mouse repertoire of the adhesion family of G-protein-coupled receptors. _Genomics_ 84, 23–33 (2004). Article
Google Scholar * Gloriam, D.E., Schioth, H.B. & Fredriksson, R. Nine new human Rhodopsin family G-protein coupled receptors: identification, sequence characterisation and evolutionary
relationship. _Biochim. Biophys. Acta_ 1722, 235–246 (2005). Article CAS Google Scholar * Cruz, M.T. et al. Type 7 adenylyl cyclase is involved in the ethanol and CRF sensitivity of
GABAergic synapses in mouse central amygdala. _Front. Neurosci._ 4, 207 (2011). Article Google Scholar * Vacic, V. et al. Duplications of the neuropeptide receptor gene _VIPR2_ confer
significant risk for schizophrenia. _Nature_ 471, 499–503 (2011). Article CAS Google Scholar * Stefansson, H. et al. Large recurrent microdeletions associated with schizophrenia. _Nature_
455, 232–236 (2008). Article CAS Google Scholar * International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. _Nature_ 455,
237–241 (2008). * Walsh, T. et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. _Science_ 320, 539–543 (2008). Article CAS Google
Scholar * Fénelon, K. et al. Deficiency of _Dgcr8_, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. _Proc. Natl. Acad. Sci.
USA_ 108, 4447–4452 (2011). Article Google Scholar * Sigurdsson, T., Stark, K.L., Karayiorgou, M., Gogos, J.A. & Gordon, J.A. Impaired hippocampal-prefrontal synchrony in a genetic
mouse model of schizophrenia. _Nature_ 464, 763–767 (2010). Article CAS Google Scholar * Arguello, P.A. & Gogos, J.A. Cognition in mouse models of schizophrenia susceptibility genes.
_Schizophr. Bull._ 36, 289–300 (2010). Article Google Scholar * Arguello, P.A. & Gogos, J.A. Modeling madness in mice: one piece at a time. _Neuron_ 52, 179–196 (2006). Article CAS
Google Scholar * Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. _Nat. Biotechnol._ 27, 182–189 (2009). Article CAS
Google Scholar Download references ACKNOWLEDGEMENTS We thank all the families who participated in this research. We also thank H. Pretorius and nursing sisters R. van Wyk, C. Botha and H.
van den Berg for their assistance with subject recruitment, family history assessments and diagnostic evaluations. We thank Y. Sun for technical assistance with DNA extractions and sample
preparations and J. Grun for information technology support. We also thank E. Fledderman and S. Thomas for support of the sequencing studies and M. Robinson for critical project support.
This work was supported in part by National Institute of Mental Health (NIMH) grants MH061399 (to M.K.) and MH077235 (to J.A.G.) and the Lieber Center for Schizophrenia Research at Columbia
University. B.X. was partially supported by a National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Award. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS *
Department of Psychiatry, Columbia University, New York, New York, USA Bin Xu & Maria Karayiorgou * Department of Physiology & Cellular Biophysics, Columbia University, New York, New
York, USA Bin Xu & Joseph A Gogos * Weskoppies Hospital & Department of Psychiatry, University of Pretoria, Pretoria, South Africa J Louw Roos * HudsonAlpha Institute for
Biotechnology, Huntsville, Alabama, USA Phillip Dexheimer, Braden Boone, Brooks Plummer & Shawn Levy * Department of Neuroscience, Columbia University, New York, New York, USA Joseph A
Gogos Authors * Bin Xu View author publications You can also search for this author inPubMed Google Scholar * J Louw Roos View author publications You can also search for this author
inPubMed Google Scholar * Phillip Dexheimer View author publications You can also search for this author inPubMed Google Scholar * Braden Boone View author publications You can also search
for this author inPubMed Google Scholar * Brooks Plummer View author publications You can also search for this author inPubMed Google Scholar * Shawn Levy View author publications You can
also search for this author inPubMed Google Scholar * Joseph A Gogos View author publications You can also search for this author inPubMed Google Scholar * Maria Karayiorgou View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS B.X., J.A.G. and M.K. designed the study, interpreted the data and prepared the manuscript. B.X.
developed the analysis pipeline and had the primary role in analysis and validation of sequence data. J.L.R. collected the samples and was the primary clinician on the project. S.L. and B.P.
performed exome library construction, capture and sequencing. P.D. contributed to the analysis of the data. B.B. contributed to the primary sequence data analysis. S.L. supervised the
sequencing project at HudsonAlpha Institute and contributed to the manuscript. CORRESPONDING AUTHORS Correspondence to Joseph A Gogos or Maria Karayiorgou. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Note, Supplementary Figures 1–4 and Supplementary
Tables 1 and 2. (PDF 2302 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Xu, B., Roos, J., Dexheimer, P. _et al._ Exome sequencing supports a _de
novo_ mutational paradigm for schizophrenia. _Nat Genet_ 43, 864–868 (2011). https://doi.org/10.1038/ng.902 Download citation * Received: 26 May 2011 * Accepted: 12 July 2011 * Published: 07
August 2011 * Issue Date: September 2011 * DOI: https://doi.org/10.1038/ng.902 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable
link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative