Systematic interpretation of genetic interactions using protein networks
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
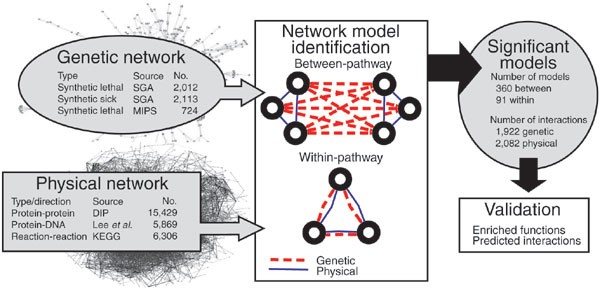
ABSTRACT Genetic interaction analysis,in which two mutations have a combined effect not exhibited by either mutation alone, is a powerful and widespread tool for establishing functional
linkages between genes. In the yeast _Saccharomyces cerevisiae_, ongoing screens have generated >4,800 such genetic interaction data. We demonstrate that by combining these data with
information on protein-protein, prote in-DNA or metabolic networks, it is possible to uncover physical mechanisms behind many of the observed genetic effects. Using a probabilistic model, we
found that 1,922 genetic interactions are significantly associated with either between- or within-pathway explanations encoded in the physical networks, covering ∼40% of known genetic
interactions. These models predict new functions for 343 proteins and suggest that between-pathway explanations are better than within-pathway explanations at interpreting genetic
interactions identified in systematic screens. This study provides a road map for how genetic and physical interactions can be integrated to reveal pathway organization and function. Access
through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal
Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices
may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support
SIMILAR CONTENT BEING VIEWED BY OTHERS ASSOCIATION STUDY BASED ON TOPOLOGICAL CONSTRAINTS OF PROTEIN–PROTEIN INTERACTION NETWORKS Article Open access 01 July 2020 GAINING CONFIDENCE IN
INFERRED NETWORKS Article Open access 14 February 2022 RESOLVING DISCREPANCIES BETWEEN CHIMERIC AND MULTIPLICATIVE MEASURES OF HIGHER-ORDER EPISTASIS Article Open access 17 February 2025
REFERENCES * Guarente, L. Synthetic enhancement in gene interaction: a genetic tool come of age. _Trends Genet._ 9, 362–366 (1993). Article CAS Google Scholar * Avery, L. & Wasserman,
S. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. _Trends Genet._ 8, 312–316 (1992). Article CAS Google Scholar * Thomas, J.H. Thinking about genetic
redundancy. _Trends Genet._ 9, 395–399 (1993). Article CAS Google Scholar * Hartman, J.L., Garvik, B. & Hartwell, L. Principles for the buffering of genetic variation. _Science_ 291,
1001–1004 (2001). Article CAS Google Scholar * Sham, P. Shifting paradigms in gene-mapping methodology for complex traits. _Pharmacogenomics_ 2, 195–202 (2001). Article CAS Google
Scholar * Dolma, S., Lessnick, S.L., Hahn, W.C. & Stockwell, B.R. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human
tumor cells. _Cancer Cell_ 3, 285–296 (2003). Article CAS Google Scholar * Forsburg, S.L. The art and design of genetic screens: yeast. _Nat. Rev. Genet._ 2, 659–668 (2001). Article CAS
Google Scholar * Tong, A.H. et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. _Science_ 294, 2364–2368 (2001). Article CAS Google Scholar * Ooi, S.L.,
Shoemaker, D.D. & Boeke, J.D. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. _Nat. Genet._ 35, 277–286 (2003). Article CAS Google
Scholar * Tong, A.H. et al. Global mapping of the yeast genetic interaction network. _Science_ 303, 808–813 (2004). Article CAS Google Scholar * Li, S. et al. A map of the interactome
network of the metazoan _C. elegans_. _Science_ 303, 540–543 (2004). Article CAS Google Scholar * Rain, J.C. et al. The protein-protein interaction map of _Helicobacter pylori_. _Nature_
409, 211–215 (2001). Article CAS Google Scholar * Uetz, P. et al. A comprehensive analysis of protein-protein interactions in _Saccharomyces cerevisiae_. _Nature_ 403, 623–627 (2000).
Article CAS Google Scholar * Giot, L. et al. A protein interaction map of _Drosophila melanogaster_. _Science_ 302, 1727–1736 (2003). Article CAS Google Scholar * Ito, T. et al. Toward
a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. _Proc. Natl. Acad.
Sci. USA_ 97, 1143–1147 (2000). Article CAS Google Scholar * Ho, Y. et al. Systematic identification of protein complexes in _Saccharomyces cerevisiae_ by mass spectrometry. _Nature_
415, 180–183 (2002). Article CAS Google Scholar * Gavin, A-C., Bösche, M., Krause, R., Grandi, P. & Marzioch, M. Functional organization of the yeast proteome by systematic analysis
of protein complexes. _Nature_ 415, 141–147 (2002). Article CAS Google Scholar * Lee, T.I. et al. Transcriptional regulatory networks in _Saccharomyces cerevisiae_. _Science_ 298, 799–804
(2002). Article CAS Google Scholar * Harbison, C.T. et al. Transcriptional regulatory code of a eukaryotic genome. _Nature_ 431, 99–104 (2004). Article CAS Google Scholar * Ozier, O.,
Amin, N. & Ideker, T. Global architecture of genetic interactions on the protein network. _Nat. Biotechnol._ 21, 490–491 (2003). Article CAS Google Scholar * Tucker, C.L. &
Fields, S. Lethal combinations. _Nat. Genet._ 35, 204–205 (2003). Article CAS Google Scholar * Winzeler, E.A. et al. Functional characterization of the _S. cerevisiae_ genome by gene
deletion and parallel analysis. _Science_ 285, 901–906 (1999). Article CAS Google Scholar * Mewes, H.W. et al. MIPS: a database for genomes and protein sequences. _Nucleic Acids Res._ 30,
31–34 (2002). Article CAS Google Scholar * Xenarios, I. et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. _Nucleic
Acids Res._ 30, 303–305 (2002). Article CAS Google Scholar * Kanehisa, M., Goto, S., Kawashima, S. & Nakaya, A. The KEGG databases at GenomeNet. _Nucleic Acids Res._ 30, 42–46
(2002). Article CAS Google Scholar * Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. _Genome Res._ 13, 2498–2504 (2003).
Article CAS Google Scholar * Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. _Genome Res._ 11, 1425–1433 (2001). * Zar, J.H. _Biostatistical
Analysis_, edn. 3 (Prentice Hall, New Jersey, 1996). Google Scholar * Kendall, S.M., Stuart, A. & Ord, J.K. _Kendall's Advanced Theory of Statistics_, edn. 5 (Oxford University
Press, NY, 1987). Google Scholar * Milo, R. et al. Network motifs: simple building blocks of complex networks. _Science_ 298, 824–827 (2002). Article CAS Google Scholar * Geissler, S.,
Siegers, K. & Schiebel, E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. _EMBO J._ 17, 952–966 (1998). Article CAS Google Scholar * Kahana, J.A.
et al. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. _Mol. Biol. Cell_ 9, 1741–1756 (1998). Article CAS
Google Scholar * Pidoux, A.L. & Allshire, R.C. Centromeres: getting a grip of chromosomes. _Curr. Opin. Cell Biol._ 12, 308–319 (2000). Article CAS Google Scholar * Pfeffer, S.R.
Membrane transport: retromer to the rescue. _Curr. Biol._ 11, R109–R111 (2001). Article CAS Google Scholar * Siniossoglou, S., Peak-Chew, S.Y. & Pelham, H.R. Ric1p and Rgp1p form a
complex that catalyses nucleotide exchange on Ypt6p. _EMBO J._ 19, 4885–4894 (2000). Article CAS Google Scholar * Hwang, W.W. et al. A conserved RING finger protein required for histone
H2B monoubiquitination and cell size control. _Mol. Cell_ 11, 261–266 (2003). Article CAS Google Scholar * Sharan, R., Ideker, T., Kelley, B.P., Shamir, R. & Karp, R.M. Identification
of protein complexes by comparative analysis of yeast and bacterial protein interaction data. _Proceedings of the Eighth Annual International Conference on Research in Computational
Molecular Biology–RECOMB_, 282–289 (ACM Press, New York, 2004) Google Scholar Download references ACKNOWLEDGEMENTS We thank Jonathan Wang, Owen Ozier and Gopal Ramachandran for preliminary
investigations and Vineet Bafna, Ben Raphael and Vikas Bansal for insightful commentary. Craig Mak, Silpa Suthram and Taylor Sittler provided helpful reviews of the text. Funding was
provided by the National Institute of General Medical Sciences (GM070743-01) and the National Science Foundation (NSF 0425926). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Program in
Bioinformatics, University of California, San Diego, 9500 Gilman Dr., San Diego, 92093-0412, California, USA Ryan Kelley & Trey Ideker * Department of Bioengineering, University of
California, San Diego, 9500 Gilman Dr., San Diego, 92093-0412, California, USA Ryan Kelley & Trey Ideker Authors * Ryan Kelley View author publications You can also search for this
author inPubMed Google Scholar * Trey Ideker View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Trey Ideker. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIG. 1 Direct overlaps between genetic and physical
interactions, while statistically significant, are limited in systematic data and probably biased. (PDF 192 kb) SUPPLEMENTARY FIG. 2 Influence of beta on result set. (PDF 179 kb)
SUPPLEMENTARY FIG. 3 Estimated prediction accuracy for naive and pathway-based within-pathway genetic predictions. (PDF 188 kb) SUPPLEMENTARY TABLE 1 Compounds excluded from the physical
interaction network (not used to connect two proteins in a metabolic interaction). (PDF 19 kb) SUPPLEMENTARY TABLE 2 The members of pathways identified in various searches. (PDF 68 kb)
SUPPLEMENTARY TABLE 3 The log-odds score associated with each network model identified in various searches. (PDF 62 kb) SUPPLEMENTARY TABLE 4 Results from reduced searches. (PDF 28 kb)
SUPPLEMENTARY TABLE 5 Functional enrichment. (PDF 23 kb) SUPPLEMENTARY TABLE 6 GO annotation predictions made with pathways obtained from various searches. (PDF 112 kb) SUPPLEMENTARY TABLE 7
Basis of annotation predictions. (PDF 21 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kelley, R., Ideker, T. Systematic interpretation of genetic
interactions using protein networks. _Nat Biotechnol_ 23, 561–566 (2005). https://doi.org/10.1038/nbt1096 Download citation * Published: 05 May 2005 * Issue Date: 01 May 2005 * DOI:
https://doi.org/10.1038/nbt1096 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative