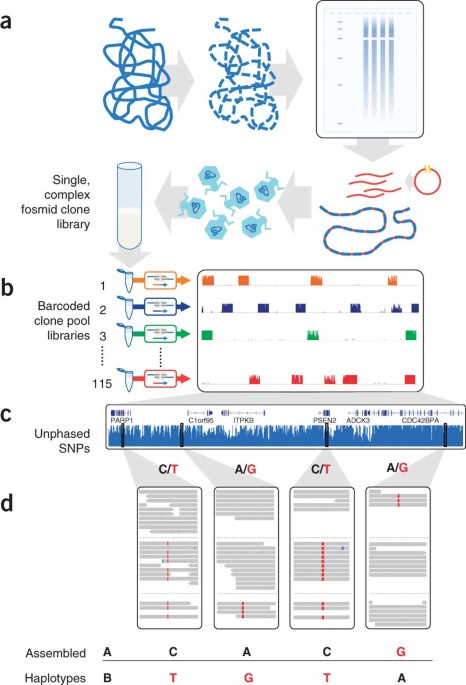
Haplotype-resolved genome sequencing of a gujarati indian individual
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Haplotype information is essential to the complete description and interpretation of genomes1, genetic diversity2 and genetic ancestry3. Although individual human genome sequencing
is increasingly routine4, nearly all such genomes are unresolved with respect to haplotype. Here we combine the throughput of massively parallel sequencing5 with the contiguity information
provided by large-insert cloning6 to experimentally determine the haplotype-resolved genome of a South Asian individual. A single fosmid library was split into a modest number of pools, each
providing ∼3% physical coverage of the diploid genome. Sequencing of each pool yielded reads overwhelmingly derived from only one homologous chromosome at any given location. These data
were combined with whole-genome shotgun sequence to directly phase 94% of ascertained heterozygous single nucleotide polymorphisms (SNPs) into long haplotype blocks (N50 of 386 kilobases
(kbp)). This method also facilitates the analysis of structural variation, for example, to anchor novel insertions7,8 to specific locations and haplotypes. Access through your institution
Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and
online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes
which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY
OTHERS FULLY PHASED HUMAN GENOME ASSEMBLY WITHOUT PARENTAL DATA USING SINGLE-CELL STRAND SEQUENCING AND LONG READS Article Open access 07 December 2020 HAPLOTYPE-RESOLVED ASSEMBLY OF DIPLOID
GENOMES WITHOUT PARENTAL DATA Article 24 March 2022 HUMAN DE NOVO MUTATION RATES FROM A FOUR-GENERATION PEDIGREE REFERENCE Article Open access 23 April 2025 ACCESSION CODES ACCESSIONS
SEQUENCE READ ARCHIVE * 026360 CHANGE HISTORY * _ 12 APRIL 2011 In the version of this supplementary file originally posted online, Supplementary Figure 4a was not properly drawn. The error
has been corrected in this file as of 12 April 2011. _ REFERENCES * Levy, S. et al. The diploid genome sequence of an individual human. _PLoS Biol._ 5, e254 (2007). Article PubMed PubMed
Central Google Scholar * International HapMap Consortium. Integrating common and rare genetic variation in diverse human populations. _Nature_ 467, 52–58 (2010). * Green, R.E. et al. A
draft sequence of the Neandertal genome. _Science_ 328, 710–722 (2010). Article CAS PubMed PubMed Central Google Scholar * Anonymous. Human genome: Genomes by the thousand. _Nature_
467, 1026–1027 (2010). * Shendure, J. & Ji, H. Next-generation DNA sequencing. _Nat. Biotechnol._ 26, 1135–1145 (2008). Article CAS PubMed Google Scholar * Kidd, J.M. et al. Mapping
and sequencing of structural variation from eight human genomes. _Nature_ 453, 56–64 (2008). CAS PubMed PubMed Central Google Scholar * Li, R. et al. Building the sequence map of the
human pan-genome. _Nat. Biotechnol._ 28, 57–63 (2010). Article CAS PubMed Google Scholar * Kidd, J.M. et al. Characterization of missing human genome sequences and copy-number
polymorphic insertions. _Nat. Methods_ 7, 365–371 (2010). Article CAS PubMed PubMed Central Google Scholar * International Human Genome Sequencing Consortium. Initial sequencing and
analysis of the human genome. _Nature_ 409, 860–921 (2001). * McKernan, K.J. et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation
sequencing using two-base encoding. _Genome Res._ 19, 1527–1541 (2009). Article CAS PubMed PubMed Central Google Scholar * Schatz, M.C., Delcher, A.L. & Salzberg, S.L. Assembly of
large genomes using second-generation sequencing. _Genome Res._ 20, 1165–1173 (2010). Article CAS PubMed PubMed Central Google Scholar * Wang, J. et al. The diploid genome sequence of
an Asian individual. _Nature_ 456, 60–65 (2008). Article CAS PubMed PubMed Central Google Scholar * Roach, J.C. et al. Analysis of genetic inheritance in a family quartet by
whole-genome sequencing. _Science_ 328, 636–639 (2010). Article CAS PubMed PubMed Central Google Scholar * 1000 Genomes Project Consortium. A map of human genome variation from
population-scale sequencing. _Nature_ 467, 1061–1073 (2010). * Reich, D., Thangaraj, K., Patterson, N., Price, A.L. & Singh, L. Reconstructing Indian population history. _Nature_ 461,
489–494 (2009). Article CAS PubMed PubMed Central Google Scholar * Adey, A. et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high density _in vitro_
transposition. _Genome Biol._ 11, R119 (2010). Article CAS PubMed PubMed Central Google Scholar * Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler
transform. _Bioinformatics_ 25, 1754–1760 (2009). Article CAS PubMed PubMed Central Google Scholar * McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing
next-generation DNA sequencing data. _Genome Res._ 20, 1297–1303 (2010). Article CAS PubMed PubMed Central Google Scholar * Bansal, V. & Bafna, V. HapCUT: an efficient and accurate
algorithm for the haplotype assembly problem. _Bioinformatics_ 24, i153–i159 (2008). Article PubMed Google Scholar * Kim, J.H., Waterman, M.S. & Li, L.M. Diploid genome reconstruction
of Ciona intestinalis and comparative analysis with Ciona savignyi. _Genome Res._ 17, 1101–1110 (2007). Article CAS PubMed PubMed Central Google Scholar * Bansal, V., Halpern, A.L.,
Axelrod, N. & Bafna, V. An MCMC algorithm for haplotype assembly from whole-genome sequence data. _Genome Res._ 18, 1336–1346 (2008). Article CAS PubMed PubMed Central Google Scholar
* Alkan, C. et al. Personalized copy number and segmental duplication maps using next-generation sequencing. _Nat. Genet._ 41, 1061–1067 (2009). Article CAS PubMed PubMed Central
Google Scholar * Hormozdiari, F. et al. Next-generation VariationHunter: combinatorial algorithms for transposon insertion discovery. _Bioinformatics_ 26, i350–i357 (2010). Article CAS
PubMed PubMed Central Google Scholar * Zody, M.C. et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. _Nat. Genet._ 40, 1076–1083 (2008). Article CAS PubMed PubMed
Central Google Scholar * Ng, S.B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. _Nature_ 461, 272–276 (2009). Article CAS PubMed PubMed Central Google
Scholar * Ng, S.B. et al. Exome sequencing identifies the cause of a mendelian disorder. _Nat. Genet._ 42, 30–35 (2010). Article CAS PubMed Google Scholar * Drysdale, C.M. et al.
Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. _Proc. Natl. Acad. Sci. USA_ 97, 10483–10488 (2000).
Article CAS PubMed PubMed Central Google Scholar * Korbel, J.O. et al. Paired-end mapping reveals extensive structural variation in the human genome. _Science_ 318, 420–426 (2007).
Article CAS PubMed PubMed Central Google Scholar * Ma, L. et al. Direct determination of molecular haplotypes by chromosome microdissection. _Nat. Methods_ 7, 299–301 (2010). Article
CAS PubMed PubMed Central Google Scholar * Tycko, B. Allele-specific DNA methylation: beyond imprinting. _Hum. Mol. Genet._ 19, R210–R220 (2010). Article CAS PubMed PubMed Central
Google Scholar * Raymond, C.K. et al. Targeted, haplotype-resolved resequencing of long segments of the human genome. _Genomics_ 86, 759–766 (2005). Article CAS PubMed Google Scholar *
Sudmant, P.H. et al. Diversity of human copy number variation and multicopy genes. _Science_ 330, 641–646 (2010). Article CAS PubMed PubMed Central Google Scholar * Zerbino, D.R. &
Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. _Genome Res._ 18, 821–829 (2008). Article CAS PubMed PubMed Central Google Scholar Download
references ACKNOWLEDGEMENTS We thank C. Lee and M. Malig for technical assistance, J. Akey, T. O'Connor and P. Green for helpful discussions, D. Reich for ancestry information on
NA20847, the U.W. Genome Sciences Genomics Resource Center (GS-GRC) for sequencing and the 1000 Genomes Project for early data release. This work was supported by National Institutes of
Health grants AG039173 (J.B.H.) and HG002385 (E.E.E.), a National Science Foundation Graduate Research Fellowship (J.O.K.), a Natural Sciences and Engineering Research Council of Canada
Fellowship (P.H.S.) and a fellowship from the Achievement Rewards for College Scientists Foundation (J.B.H.). E.E.E. is an investigator of the Howard Hughes Medical Institute. AUTHOR
INFORMATION AUTHORS AND AFFILIATIONS * Department of Genome Sciences, University of Washington School of Medicine, Seattle, Washington, USA Jacob O Kitzman, Alexandra P MacKenzie, Andrew
Adey, Joseph B Hiatt, Rupali P Patwardhan, Peter H Sudmant, Sarah B Ng, Can Alkan, Ruolan Qiu, Evan E Eichler & Jay Shendure * Howard Hughes Medical Institute, Seattle, Washington, USA
Can Alkan & Evan E Eichler Authors * Jacob O Kitzman View author publications You can also search for this author inPubMed Google Scholar * Alexandra P MacKenzie View author publications
You can also search for this author inPubMed Google Scholar * Andrew Adey View author publications You can also search for this author inPubMed Google Scholar * Joseph B Hiatt View author
publications You can also search for this author inPubMed Google Scholar * Rupali P Patwardhan View author publications You can also search for this author inPubMed Google Scholar * Peter H
Sudmant View author publications You can also search for this author inPubMed Google Scholar * Sarah B Ng View author publications You can also search for this author inPubMed Google Scholar
* Can Alkan View author publications You can also search for this author inPubMed Google Scholar * Ruolan Qiu View author publications You can also search for this author inPubMed Google
Scholar * Evan E Eichler View author publications You can also search for this author inPubMed Google Scholar * Jay Shendure View author publications You can also search for this author
inPubMed Google Scholar CONTRIBUTIONS The project was conceived and experiments planned by J.O.K., E.E.E. and J.S. J.O.K., A.P.M. and R.Q. carried out all experiments. J.O.K., A.A., J.B.H.,
R.P.P., P.H.S., S.B.N. and C.A. performed data analysis. J.O.K., A.P.M., A.A., J.B.H., R.P.P. and J.S. wrote the manuscript, and all authors reviewed it. All aspects of the study were
supervised by J.S. CORRESPONDING AUTHORS Correspondence to Jacob O Kitzman or Jay Shendure. ETHICS DECLARATIONS COMPETING INTERESTS J.S. is a member of the science advisory boards of Tandem
Technologies, Stratos Genomics, Good Start Genetics and Adaptive TCR. E.E.E. is on the scientific advisory board for Pacific Biosciences. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND
FIGURES Supplementary Tables 1–3,5, Supplementary Methods and Supplementary Figs. 1–7 (PDF 1756 kb) SUPPLEMENTARY TABLE 4 Pan-genome and novel sequence anchoring. (XLS 1322 kb) RIGHTS AND
PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kitzman, J., MacKenzie, A., Adey, A. _et al._ Haplotype-resolved genome sequencing of a Gujarati Indian individual.
_Nat Biotechnol_ 29, 59–63 (2011). https://doi.org/10.1038/nbt.1740 Download citation * Received: 26 October 2010 * Accepted: 29 November 2010 * Published: 01 January 2011 * Issue Date:
January 2011 * DOI: https://doi.org/10.1038/nbt.1740 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link
is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative