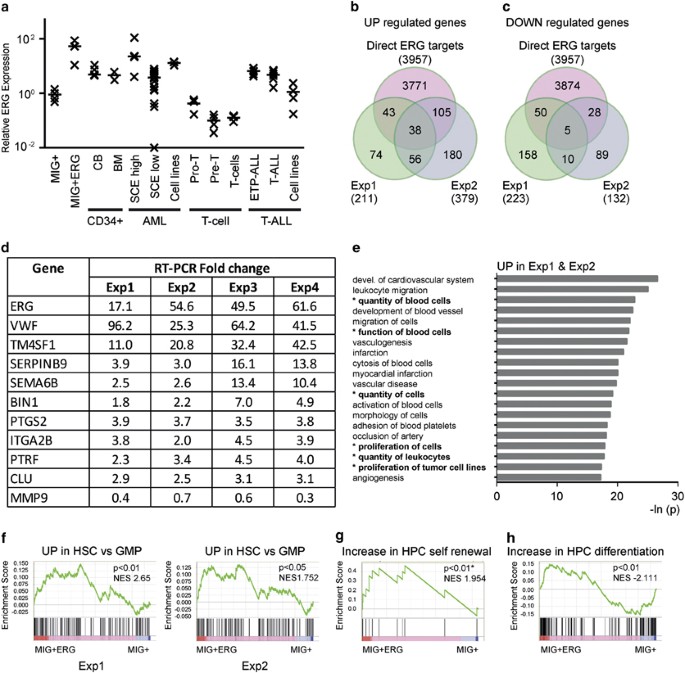
Overexpression of erg in cord blood progenitors promotes expansion and recapitulates molecular signatures of high erg leukemias
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT High expression of the ETS family transcription factor ERG is associated with poor clinical outcome in acute myeloid leukemia (AML) and acute T-cell lymphoblastic leukemia (T-ALL).
In murine models, high ERG expression induces both T-ALL and AML. However, no study to date has defined the effect of high ERG expression on primary human hematopoietic cells. In the present
study, human CD34+ cells were transduced with retroviral vectors to elevate ERG gene expression to levels detected in high ERG AML. RNA sequencing was performed on purified populations of
transduced cells to define the effects of high ERG on gene expression in human CD34+ cells. Integration of the genome-wide expression data with other data sets revealed that high ERG drives
an expression signature that shares features of normal hematopoietic stem cells, high ERG AMLs, early T-cell precursor-ALLs and leukemic stem cell signatures associated with poor clinical
outcome. Functional assays linked this gene expression profile to enhanced progenitor cell expansion. These results support a model whereby a stem cell gene expression network driven by high
ERG in human cells enhances the expansion of the progenitor pool, providing opportunity for the acquisition and propagation of mutations and the development of leukemia. Access through your
institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print
issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to
local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT
BEING VIEWED BY OTHERS INCREASED EXPRESSION OF RUNX3 INHIBITS NORMAL HUMAN MYELOID DEVELOPMENT Article Open access 30 April 2022 HMX3 IS A CRITICAL VULNERABILITY IN MECOM-NEGATIVE
KMT2A::MLLT3 ACUTE MYELOMONOCYTIC LEUKEMIA Article 04 December 2024 SINGLE-CELL RNA SEQUENCING OF A NEW TRANSGENIC T(8;21) PRELEUKEMIA MOUSE MODEL REVEALS REGULATORY NETWORKS PROMOTING
LEUKEMIC TRANSFORMATION Article Open access 14 October 2023 REFERENCES * Beck D, Thoms JA, Perera D, Schutte J, Unnikrishnan A, Knezevic K _et al_. Genome-wide analysis of transcriptional
regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. _Blood_ 2013; 122: e12–e22. Article CAS Google Scholar * Wilson NK, Foster SD, Wang X,
Knezevic K, Schutte J, Kaimakis P _et al_. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. _Cell Stem
Cell_ 2010; 7: 532–544. Article CAS Google Scholar * Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA _et al_. The transcription factor Erg is essential for
definitive hematopoiesis and the function of adult hematopoietic stem cells. _Nat Immunol_ 2008; 9: 810–819. Article CAS Google Scholar * Ng AP, Loughran SJ, Metcalf D, Hyland CD, de
Graaf CA, Hu Y _et al_. Erg is required for self-renewal of hematopoietic stem cells during stress hematopoiesis in mice. _Blood_ 2011; 118: 2454–2461. Article CAS Google Scholar *
Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME _et al_. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. _Cell_ 2011;
144: 296–309. Article CAS Google Scholar * Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV . Precise developmental regulation of Ets family transcription factors during
specification and commitment to the T cell lineage. _Development_ 1999; 126: 3131–3148. CAS Google Scholar * Baldus CD, Burmeister T, Martus P, Schwartz S, Gokbuget N, Bloomfield CD _et
al_. High expression of the ETS transcription factor ERG predicts adverse outcome in acute T-lymphoblastic leukemia in adults. _J Clin Oncol_ 2006; 24: 4714–4720. Article CAS Google
Scholar * Metzeler KH, Dufour A, Benthaus T, Hummel M, Sauerland MC, Heinecke A _et al_. ERG expression is an independent prognostic factor and allows refined risk stratification in
cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. _J Clin Oncol_ 2009; 27: 5031–5038.
Article CAS Google Scholar * Marcucci G, Maharry K, Whitman SP, Vukosavljevic T, Paschka P, Langer C _et al_. High expression levels of the ETS-related gene, ERG, predict adverse outcome
and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B Study. _J Clin Oncol_ 2007; 25: 3337–3343. Article CAS
Google Scholar * Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P _et al_. Stem cell gene expression programs influence clinical outcome in human leukemia. _Nat Med_
2011; 17: 1086–1093. Article CAS Google Scholar * Diffner E, Beck D, Gudgin E, Thoms JA, Knezevic K, Pridans C _et al_. Activity of a heptad of transcription factors is associated with
stem cell programs and clinical outcome in acute myeloid leukemia. _Blood_ 2013; 121: 2289–2300. Article CAS Google Scholar * Thoms JA, Birger Y, Foster S, Knezevic K, Kirschenbaum Y,
Chandrakanthan V _et al_. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. _Blood_ 2011; 117: 7079–7089. Article CAS
Google Scholar * Salek-Ardakani S, Smooha G, de Boer J, Sebire NJ, Morrow M, Rainis L _et al_. ERG is a megakaryocytic oncogene. _Cancer Res_ 2009; 69: 4665–4673. Article CAS Google
Scholar * Goldberg L, Tijssen MR, Birger Y, Hannah RL, Kinston SJ, Schutte J _et al_. Genome-scale expression and transcription factor binding profiles reveal therapeutic targets in
transgenic ERG myeloid leukemia. _Blood_ 2013; 122: 2694–2703. Article CAS Google Scholar * Tsuzuki S, Taguchi O, Seto M . Promotion and maintenance of leukemia by ERG. _Blood_ 2011; 117:
3858–3868. Article CAS Google Scholar * Carmichael CL, Metcalf D, Henley KJ, Kruse EA, Di Rago L, Mifsud S _et al_. Hematopoietic overexpression of the transcription factor Erg induces
lymphoid and erythro-megakaryocytic leukemia. _Proc Natl Acad Sci USA_ 2012; 109: 15437–15442. Article CAS Google Scholar * Pereira DS, Dorrell C, Ito CY, Gan OI, Murdoch B, Rao VN _et
al_. Retroviral transduction of TLS-ERG initiates a leukemogenic program in normal human hematopoietic cells. _Proc Natl Acad Sci USA_ 1998; 95: 8239–8244. Article CAS Google Scholar *
Warner JK, Wang JC, Takenaka K, Doulatov S, McKenzie JL, Harrington L _et al_. Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic
cells. _Leukemia_ 2005; 19: 1794–1805. Article CAS Google Scholar * Rabbitts TH, Forster A, Larson R, Nathan P . Fusion of the dominant negative transcription regulator CHOP with a novel
gene FUS by translocation t(12;16) in malignant liposarcoma. _Nat Genet_ 1993; 4: 175–180. Article CAS Google Scholar * Cheng L, Fu J, Tsukamoto A, Hawley RG . Use of green fluorescent
protein variants to monitor gene transfer and expression in mammalian cells. _Nat Biotechnol_ 1996; 14: 606–609. Article CAS Google Scholar * Miller AD, Garcia JV, von Suhr N, Lynch CM,
Wilson C, Eiden MV . Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. _J Virol_ 1991; 65: 2220–2224. CAS PubMed PubMed Central Google Scholar
* Shen SW, Dolnikov A, Passioura T, Millington M, Wotherspoon S, Rice A _et al_. Mutant N-ras preferentially drives human CD34+ hematopoietic progenitor cells into myeloid differentiation
and proliferation both _in vitro_ and in the NOD/SCID mouse. _Exp Hematol_ 2004; 32: 852–860. Article CAS Google Scholar * Mulloy JC, Cammenga J, MacKenzie KL, Berguido FJ, Moore MA,
Nimer SD . The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. _Blood_ 2002; 99: 15–23. Article CAS Google Scholar * Schuller CE, Jankowski K, Mackenzie
KL . Telomere length of cord blood-derived CD34(+) progenitors predicts erythroid proliferative potential. _Leukemia_ 2007; 21: 983–991. Article CAS Google Scholar * Breems DA, Blokland
EA, Neben S, Ploemacher RE . Frequency analysis of human primitive haematopoietic stem cell subsets using a cobblestone area forming cell assay. _Leukemia_ 1994; 8: 1095–1104. CAS PubMed
Google Scholar * Holmes R, Zuniga-Pflucker JC . The OP9-DL1 system: generation of T-lymphocytes from embryonic or hematopoietic stem cells _in vitro_. _Cold Spring Harb Protoc_ 2009; 2009:
pdb prot5156 Article Google Scholar * Trapnell C, Pachter L, Salzberg SL . TopHat: discovering splice junctions with RNA-Seq. _Bioinformatics_ 2009; 25: 1105–1111. Article CAS Google
Scholar * Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P _et al_. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for
macrophage and B cell identities. _Mol Cell_ 2010; 38: 576–589. Article CAS Google Scholar * Anders S, Huber W . Differential expression analysis for sequence count data. _Genome Biol_
2010; 11: R106. Article CAS Google Scholar * Awong G, Herer E, Surh CD, Dick JE, La Motte-Mohs RN, Zuniga-Pflucker JC . Characterization _in vitro_ and engraftment potential _in vivo_ of
human progenitor T cells generated from hematopoietic stem cells. _Blood_ 2009; 114: 972–982. Article CAS Google Scholar * Spits H . Development of alphabeta T cells in the human thymus.
_Nat Rev Immunol_ 2002; 2: 760–772. Article CAS Google Scholar * Rockova V, Abbas S, Wouters BJ, Erpelinck CA, Beverloo HB, Delwel R _et al_. Risk stratification of intermediate-risk
acute myeloid leukemia: integrative analysis of a multitude of gene mutation and gene expression markers. _Blood_ 2011; 118: 1069–1076. Article CAS Google Scholar * Metzeler KH, Hummel M,
Bloomfield CD, Spiekermann K, Braess J, Sauerland MC _et al_. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. _Blood_ 2008;
112: 4193–4201. Article CAS Google Scholar * The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. _N Engl J Med_ 2013; 368:
2059–2074 Article Google Scholar * Verhaak RG, Wouters BJ, Erpelinck CA, Abbas S, Beverloo HB, Lugthart S _et al_. Prediction of molecular subtypes in acute myeloid leukemia based on gene
expression profiling. _Haematologica_ 2009; 94: 131–134. Article Google Scholar * Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA _et al_. Gene set enrichment
analysis: a knowledge-based approach for interpreting genome-wide expression profiles. _Proc Natl Acad Sci USA_ 2005; 102: 15545–15550. Article CAS Google Scholar * Zhang J, Ding L,
Holmfeldt L, Wu G, Heatley SL, Payne-Turner D _et al_. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. _Nature_ 2012; 481: 157–163. Article CAS Google Scholar *
Neumann M, Heesch S, Schlee C, Schwartz S, Gokbuget N, Hoelzer D _et al_. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. _Blood_ 2013; 121: 4749–4752.
Article CAS Google Scholar * Chacon D, Beck D, Perera D, Wong JW, Pimanda JE . BloodChIP: a database of comparative genome-wide transcription factor binding profiles in human blood cells.
_Nucleic Acids Res_ 2013; 42: D172–D177. Article Google Scholar * Beck D, Thoms JA, Perera D, Schutte J, Unnikrishnan A, Knezevic K _et al_. Genome-wide analysis of transcriptional
regulators in human HSPCs reveals a densely interconnected network of coding and non-coding genes. _Blood_ 2013; 122: e12–e22. Article CAS Google Scholar * Feuring-Buske M, Gerhard B,
Cashman J, Humphries RK, Eaves CJ, Hogge DE . Improved engraftment of human acute myeloid leukemia progenitor cells in beta 2-microglobulin-deficient NOD/SCID mice and in NOD/SCID mice
transgenic for human growth factors. _Leukemia_ 2003; 17: 760–763. Article CAS Google Scholar * Wunderlich M, Chou FS, Link KA, Mizukawa B, Perry RL, Carroll M _et al_. AML xenograft
efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. _Leukemia_ 2010; 24: 1785–1788. Article CAS Google Scholar * Nicolini FE,
Cashman JD, Hogge DE, Humphries RK, Eaves CJ . NOD/SCID mice engineered to express human IL-3, GM-CSF and Steel factor constitutively mobilize engrafted human progenitors and compromise
human stem cell regeneration. _Leukemia_ 2004; 18: 341–347. Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank Gillian Rozenberg for the photographs of the
cytospins, donors and staff of the CB banks at the Prince of Wales and Royal North Shore Hospitals for CBs, Simon Downes, Soo Min Heng and Joel Poder from Medical Physics, Department of
Radiation Oncology, Prince of Wales Hospital for calibration of the bio-irradiator. This work was funded by the National Health and Medical Research Council (Australia), Leukaemia Foundation
(Australia) and the Cancer Institute of NSW. MLT received scholarships from the Prince of Wales Clinical School and the Children’s Cancer Institute, Australia. Children’s Cancer Institute
is affiliated with University of New South Wales and Sydney Children’s Hospital Network. The authors also thank the Australian Research Council (JWHW), Children with Cancer (UK), Israel
Science Foundation and Waxman Cancer Research Foundation (to SI). This work was funded by the National Health and Medical Research Council (Australia) and Leukaemia Foundation (Australia).
AUTHOR CONTRIBUTIONS MLT, DB, JAIT, YH, AK, AU, KK, KE, LAR, EL and LG performed experiments and analyzed data. JO, KLM, JEP and JWHW analyzed data, JM and RBL provided essential reagents,
MLT, DB, JAIT, SI, KLM and JEP wrote the paper. AUTHOR INFORMATION Author notes * M L Tursky, D Beck, J A I Thoms, K L MacKenzie and J E Pimanda: These authors contributed equally as first
authors. * These authors contributed equally as senior authors. AUTHORS AND AFFILIATIONS * Adult Cancer Program, Prince of Wales Clinical School, Lowy Cancer Research Centre, UNSW, Sydney,
Australia M L Tursky, D Beck, J A I Thoms, Y Huang, A Unnikrishnan, K Knezevic, J W H Wong & J E Pimanda * Children’s Cancer Institute Australia, Lowy Cancer Research Centre, UNSW,
Sydney, Australia M L Tursky, A Kumari, K Evans, L A Richards, E Lee, R B Lock & K L MacKenzie * Department of Obstetrics and Gynaecology, Royal North Shore Hospital, University of
Sydney, Sydney, Australia J Morris * Cancer Research Center, Sheba Medical Center, Tel Hashomer, Ramat Gan, Israel L Goldberg & S Izraeli * Department of Human Molecular Genetics and
Biochemistry, Tel Aviv University, Tel Aviv, Israel L Goldberg & S Izraeli * School of Mathematics and Statistics, UNSW, Sydney, Australia J Olivier * Department of Haematology, Prince
of Wales Hospital, Sydney, Australia J E Pimanda Authors * M L Tursky View author publications You can also search for this author inPubMed Google Scholar * D Beck View author publications
You can also search for this author inPubMed Google Scholar * J A I Thoms View author publications You can also search for this author inPubMed Google Scholar * Y Huang View author
publications You can also search for this author inPubMed Google Scholar * A Kumari View author publications You can also search for this author inPubMed Google Scholar * A Unnikrishnan View
author publications You can also search for this author inPubMed Google Scholar * K Knezevic View author publications You can also search for this author inPubMed Google Scholar * K Evans
View author publications You can also search for this author inPubMed Google Scholar * L A Richards View author publications You can also search for this author inPubMed Google Scholar * E
Lee View author publications You can also search for this author inPubMed Google Scholar * J Morris View author publications You can also search for this author inPubMed Google Scholar * L
Goldberg View author publications You can also search for this author inPubMed Google Scholar * S Izraeli View author publications You can also search for this author inPubMed Google Scholar
* J W H Wong View author publications You can also search for this author inPubMed Google Scholar * J Olivier View author publications You can also search for this author inPubMed Google
Scholar * R B Lock View author publications You can also search for this author inPubMed Google Scholar * K L MacKenzie View author publications You can also search for this author inPubMed
Google Scholar * J E Pimanda View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to K L MacKenzie or J E Pimanda. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary Information accompanies this paper on the Leukemia website SUPPLEMENTARY
INFORMATION SUPPLEMENTARY FIGURES (DOCX 1704 KB) SUPPLEMENTARY TABLES (XLSX 116 KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Tursky, M., Beck, D.,
Thoms, J. _et al._ Overexpression of ERG in cord blood progenitors promotes expansion and recapitulates molecular signatures of high ERG leukemias. _Leukemia_ 29, 819–827 (2015).
https://doi.org/10.1038/leu.2014.299 Download citation * Received: 21 May 2014 * Revised: 04 September 2014 * Accepted: 03 October 2014 * Published: 13 October 2014 * Issue Date: April 2015
* DOI: https://doi.org/10.1038/leu.2014.299 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative