Human chit1 gene distribution: new data from mediterranean and european populations
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT A 24 bp duplication in the _CHIT1_ gene (H allele) is associated with a deficiency in the activity of chitotriosidase, an enzyme with the capability to hydrolyse chitin. A recent
study in European and two sub-Saharan populations suggested a relationship between the presence of the mutation, improved environmental conditions, and the disappearance of parasitic
diseases, including _Plasmodium falciparum_ malaria. This result was not supported by the high frequency of the 24 bp duplication in a sample from Taiwan, an area with high malaria
endemicity until 40 years ago. In this study, we analysed the frequency variability of the H allele in Mediterranean populations and its internal variability in Sardinia (Italy) with respect
to malaria, which had been endemic on the island until its eradication during 1946–1950. The pattern of H frequency distributions is not consistent with the hypothesis of selective
pressures acting on _CHIT1_ gene. The Moran’s index coefficient and correlogram seem to indicate, indeed, that allele distribution was determined by random factors. The pattern of frequency
distribution suggests a possible Asiatic origin of the H allele, but it could be possible also that the mutant allele had diffused out of Africa, and was subsequently lost from African
populations. SIMILAR CONTENT BEING VIEWED BY OTHERS _PLASMODIUM MALARIAE_ STRUCTURE AND GENETIC DIVERSITY IN SUB-SAHARAN AFRICA DETERMINED FROM MICROSATELLITE VARIANTS AND LINKED SNPS IN
ORTHOLOGUES OF ANTIMALARIAL RESISTANCE GENES Article Open access 19 December 2022 FINE SCALE HUMAN GENETIC STRUCTURE IN THREE REGIONS OF CAMEROON REVEALS EPISODIC DIVERSIFYING SELECTION
Article Open access 13 January 2021 DIVERSITY AND EVOLUTION OF THE MHC CLASS II _DRB_ GENE IN THE _CAPRA SIBIRICA_ EXPERIENCED A DEMOGRAPHIC FLUCTUATION IN CHINA Article Open access 07
November 2023 INTRODUCTION Human chitotriosidase is an enzyme synthesized by activated macrophages and has the capability to hydrolyse chitin, a structural component present in the coatings
of many living species, including fungi, parasitic nematodes, and insects. Chitotriosidase is secreted mainly as an active 50 kDa enzyme containing a C-terminal chitin-binding domain. It is
proteolytically processed to a C-terminally truncated 39 kDa isoform characterized by hydrolase activity, which accumulates in lysosomes (Renkema et al. 1997). The 50 kDa form is synthesized
by neutrophilic granulocyte progenitors and stored in their granules (Boot et al. 1995). It is considered as a component of the innate immunity that may play a role in defence against
chitin-containing pathogens. Chitotriosidase exerts activity towards chitin-containing pathogens, such as _C. neoformans_, _M. rouxi_ and _C. albicans_ both in vitro and in vivo (van Eijk et
al. 2005; Boot et al. 2001). Additional evidence of a role for chitotriosidase during immunological responses is the observation that the enzyme is shortly and acutely up-regulated, both at
the level of RNA and in activity following stimulation with prolactin, IFN-γ, TNFα, LPS, and IL-4 (Di Rosa et al. 2005; Malaguarnera et al. 2004; van Eijk et al. 2005). Moreover, _CHIT_
activity has been used in the screening of lysosomal storage diseases (Grosso et al. 2004) and in the monitoring of the treatment of Gaucher disease (Cabrera-Salazar et al. 2004). The 24 bp
duplication has been associated with susceptibility to infection by _Wuchereria bancrofti_ in south India (Choi et al. 2001) but not in Papua New Guinea (Hise et al. 2003). The discovery of
the existence of a second chitinase, named acidic mammalian chitinase (AMCase), in humans, has opened the possibility that a deficiency in chitotriosidase might be partly compensated for by
the presence of the latter enzyme (Boot et al. 2001). A significant increase in plasma chitotriosidase levels, with respect to healthy subjects, in African children infected with acute
_Plasmodium falciparum_ malaria has been reported. Moreover, chitotriosidase levels are higher in healthy African samples than in those of Caucasians (Barone et al. 2003). Increased levels
of plasma chitotriosidase have also been reported in patients with Gaucher disease (Hollak et al. 1994), in various lysosomal storage disorders, in several haematological and infectious
diseases involving activated macrophages (Guo et al. 1995; Den Tandt and Van Hoof 1996), and in patients with β-thalassaemia (Altarescu et al. 2002; Barone et al. 1999, 2002, 2001). The
_CHIT1_ gene is located on chromosome 1q31–32 and consists of 12 exons, spanning approximately 20 kb (Boot et al. 1998). A 24 bp duplication in exon 10 of the gene causes the deletion of
amino acids 344–372, resulting in a deficiency in enzyme activity. The enzyme is totally inactive in individuals homozygous for the duplication (Boot et al. 1998; Canudas et al. 2001). This
mutation is not found in anthropomorphic primates, suggesting that it originated during human evolution (Gianfrancesco and Musumeci 2004). A recent study (Malaguarnera et al. 2003) of the 24
bp duplication in the _CHIT1_ gene in some European populations suggested a relationship between the presence of the mutation, improved environmental conditions, and the disappearance of
parasitic diseases, including _P. falciparum_ malaria. Furthermore, widespread parasitic diseases and the poor social status of the area may have contributed to the maintenance of the
wild-type (wt) _CHIT1_ gene in sub-Saharan populations (Malaguarnera et al. 2003). This result was not supported by the recent study by Chien et al. (2005), in which a frequency of 58% was
reported for the 24 bp duplication, together with high frequencies of both thalassaemia and glucose 6-phosphate dehydrogenase (G6PD) deficiency, in a sample of Chinese Han individuals from
Taiwan, an island characterized by high malaria endemicity until 40 years ago (Lin et al. 1991). In the present study the _CHIT1_ gene distribution in eight Mediterranean and European
populations was analysed with the aim of increasing the knowledge of the frequency distribution of the _CHIT_ gene from a population point of view, since six of the populations under
scrutiny have not been examined for this marker until now. The relationship between _CHIT1_ and malaria was particularly investigated in the island of Sardinia, where malaria was endemic
until its eradication during 1946–1950 (Logan 1957). The aim of this study was to increase our knowledge of the distribution of the _CHIT1_ alleles in the Mediterranean area and to interpret
its variability. MATERIALS AND METHODS Samples from 1,104 individuals from continental Italy (_n_ = 99), Sardinia (Italy) (_n_ = 335), Spain (_n_ = 103), Basque country (_n_ = 60),
continental France (_n_ = 128), Corsica (France) (_n_ = 194), Turkey (_n_ = 95), and Morocco (_n_ = 90) were genotyped. The samples were from unrelated individuals of both genders, born and
resident in their countries of origin, as their relatives had been for at least three generations. Basque, Corsican, and Sardinian samples were analysed separately from Spanish, French, and
Italian samples, because of their genetic peculiarities (Calafell and Bertranpetit 1994; Varesi et al. 2000; Vona 1997). The Sardinian samples, taken from several historical–geographical
areas of the island (Sulcis, Trexenta, Campidano of Oristano, Nuorese, and Gallura), were subdivided into three altimetric zones, characterized by different past malarial endemy (Fermi 1934,
1938; Brown 1981). The protocols and procedures used in this research were undertaken in compliance with the declaration of Helsinki. DNA was extracted with the standard phenol–chloroform
technique, and polymerase chain reaction (PCR) was performed using the primers described by Hise et al. (2003) under the following conditions: 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C,
with a final extension of 5 min at 72°C. The two fragments of 99 bp and 75 bp were separated on 3% agarose gel stained with ethidium bromide. Allele frequencies were determined by direct
gene counting. Hardy–Weinberg equilibrium was tested with GENEPOP version 3.4 software (Raymond and Rousset 1995). Genic and genotypic differentiation among samples was tested with the
chi-square test. We also tested the associations between allele frequencies, latitude, and longitude with the Pearson’s correlation. The analysis was performed with SPSS version 8.0
software. The effects of natural selection on the _CHIT1_ gene were evaluated with the Ewens–Watterson neutrality test (Ewens 1972; Watterson 1978), implemented by Slatkin (1994), and was
carried out using Pypop software (version 0.6.0) (Lancaster et al. 2003). To test the spatial distribution of allele frequencies, we estimated Moran’s index (_I_), a product–moment
correlation coefficient (Cliff and Ord 1973), and one-dimensional correlograms (Sokal and Oden 1978; Sokal et al. 1989), using SAAP version 4.3 software (Wartenberg 1989). The plot of
autocorrelation coefficient _I_ against distance is referred to as a correlogram, the overall significance of which was assessed with the Bonferroni test. To infer evolutionary patterns from
correlograms, we applied the classification suggested by Barbujani (2000). RESULTS Genotype and allele frequencies of the studied Mediterranean populations are shown in Table 1. All samples
are in Hardy–Weinberg equilibrium. The frequencies of the H allele ranged from 10.5% (Morocco) to 25.0% (France). The genotype frequencies (Table 1) indicated an absence of H/H homozygotes
in two of the populations studied (Basque and Morocco). In the other populations, the frequencies of H/H homozygotes ranged from 1% (Corsica) to 9.3% (continental France). The frequencies of
wt/H heterozygotes ranged from 20.0% (Morocco) to 34.3% (continental Italy). The chi-square test showed a high variability in the _CHIT1_ allele distribution among the eight populations
(_P_ = 0.001) and in pairwise comparisons. The French sample had the greatest number of significant pairwise comparisons, with Basque country, Corsica, Sardinia, and Morocco, followed by the
Spanish sample with Basque country, Corsica, Morocco and Morocco with Italy, France and Spain. The frequencies of the H allele in these populations were very different from those in both
the sub-Saharan African populations, characterized by an absence of the 24 bp duplication, and the Asiatic population, characterized by high frequencies of the duplication (58% in Taiwan
island) (Chien et al. 2005) (Table 2). To analyse the geographic differentiation of _CHIT1_ within the Mediterranean area, we performed a spatial autocorrelation analysis with all our
samples and samples from Portugal (Rodrigues et al. 2004), Sicily (Malaguarnera et al. 2003), and Israeli (Boot et al. 1998) (Table 2). The resulting correlogram (not shown) indicated a
random spatial distribution of variation (_P_ = 0.173) with a lack of clinal variation and with no significant Moran’s _I_ coefficients for any distance classes. Pearson’s correlation did
not suggest any association between _CHIT1_ and either longitude or latitude (_P_ long = 0.356; _P_ lat = 0.900). When we considered all the 18 populations of Table 2 (Boot et al. 1998;
Malaguarnera et al. 2003; Choi et al. 2005, 2001; Hise et al. 2003), we obtained a significant _P_ value of Pearson’s coefficient for longitude (_P_ = 0.038) but not for latitude (_P_ =
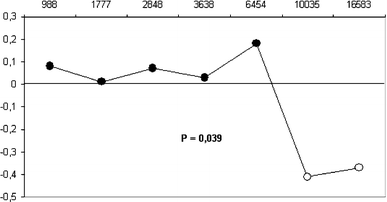
0.992). A spatial autocorrelation analysis was performed using the frequencies in Table 2. A correlogram of Moran’s index for seven distance classes showed positive but not significant
values until the 6,500 km class, beyond which Moran’s index values became negative and significant (Fig. 1). Such a pattern is partly similar to that for long-distance differentiation (LDD)
(Barbujani 2000). In fact, LDD is characterized by positive and significant values in the smallest distance classes and by negative and significant values in the largest distance classes.
The pattern is marginally significant with the Bonferroni test (_P_ = 0.039), and this may be the result of the discontinuous distribution of the population samples. The hypothesis of
selective neutrality on _CHIT1_ gene was tested with the Ewens–Watterson neutrality test (Ewens 1972; Watterson 1978; Slatkin 1994). Results suggest that all the populations do not
significantly shift from the infinite allele model. A particular analysis was performed for the island of Sardinia, which was characterized until 1945 by endemic malaria. In the Sardinian
population, the average frequency of the H allele was 17.5%, whereas the genotype frequencies were 3.5, 27.9, and 68.6 for H/H, wt/H, and wt/wt, respectively (Table 1). A comparison of our
results with previous data for the north Sardinian population (Malaguarnera et al. 2003) showed no significant difference (χ2 = 0.738; _df_ = 1; _P_ = 0.390). The Sardinian sample was
subdivided according to the municipality of origin, into three different altimetric zones: 0–200 m, 201–400 m, and >400 m. The genotype and allele frequencies are summarized in Table 3.
The frequency of the H allele decreases with altitude from 24.6% to 11.4%, as do the H/H and wt/H genotypes (from 6.3% to 0.7% and from 36.7% to 21.3%, respectively). The frequency of the
wt/wt homozygote increases with altitude, ranging from 57.0% to 77.9%. The chi-square test showed a significant degree of global differentiation among the altimetric zones (_P_ = 0.0037),
particularly at 0–200 m and >400 m (_P_ < 0.001). We recalculated the frequencies quoted by Sanna et al. (1997) for G6PD deficiency in 103 municipalities and by Siniscalco et al.
(1966) for thalassaemia in 52 municipalities, grouping the data in the altimetric zones used in our study (Table 4). The allele frequencies for G6PD deficiency decrease regularly with
increasing altitude, and thalassaemia frequencies are lower at altitudes above 400 m, whereas they are similar in the other two altimetric zones. A χ2 test showed significant differences
among altitude zones for both traits. The wt/H and H/H genotype frequencies for the _CHIT1_ locus and the frequency of the H allele decrease with altitude, as do the frequencies of the
thalassaemia and G6PD deficiency alleles. The contrary trend was followed by wt/wt genotype and wt allele frequencies. DISCUSSION A recent study of African and Mediterranean populations
(Malaguarnera et al. 2003) suggested that subjects carrying the mutant H allele at the _CHIT1_ locus have increased susceptibility to parasitic diseases. The persistence of parasitic
diseases in sub-Saharan Africa would be favoured by the maintenance of the wild-type allele, as demonstrated by the low incidence of heterozygotes wt/H and by the absence of homozygotes H/H
in these regions. Therefore, Malaguarnera et al. (2003) suggested that low frequencies of the H allele represent a protective factor in populations living under environmental conditions
favourable to parasitic diseases, such as malaria, since the individuals bearing the H allele might exhibit elevated susceptibility to infective diseases. Our analysis of samples from the
island of Sardinia does not confirm the suggestion made by these authors. In fact, the highest frequency of the H allele appeared in the lowlands, where the incidence of malaria was highest.
On the other hand, we found the highest frequency of the wt allele at altitudes over 400 m, in villages with the lowest or negligible endemic malaria (Fermi 1934, 1938). In Sardinia, the
frequency pattern of the wt allele varies with altitude in a direction opposite to that of G6PD deficiency (GdMed) and thalassaemia (Th), which are correlated with malaria (Siniscalco et al.
1966). Analysis of sample β039 carriers from Sardinia and Corsica does not show significant differences for genotype and allele frequencies with respect to healthy individuals (Piras et al.
2006). Our results seem to confirm the data reported by Chien et al. (2005) for Taiwan, where the frequency of the H allele is very high (58%), even though malaria was eradicated only 40
years ago, and those reported by Choi et al. (2001) for south India, where malaria is still present, and the frequency of the mutant allele is 44%. The distribution of the _CHIT1_ gene was
also analysed in eight Mediterranean populations. The results highlight significant heterogeneity among the populations studied. Moreover, the frequencies appear remarkably different from
those of African and Asian populations. The variability of the _CHIT1_ gene frequencies in the Mediterranean area does not appear to be linked to its spatial distribution. In fact, the
correlation of gene frequencies with latitude and longitude is not statistically significant, and the Moran index and Bonferroni’s test for the correlogram support this result. The
correlogram is evidence for the random distribution of the _CHIT1_ allele frequencies in these Mediterranean populations. The spatial distribution of _CHIT1_ allele frequencies at the
microgeographic and macrogeographic levels does not provide evidence of spatial patterns that can be interpreted as selection effects, which in many populations have produced the high H
allele frequencies associated with thalassaemia, haemoglobin variants, and G6PD deficiency. The Moran’s index coefficients and correlogram suggest that these allele distributions were
determined by random factors. This result is confirmed by the Ewens–Watterson neutrality test, which suggests the absence of natural selection on the _CHIT1_ gene. This analysis of
populations in the Mediterranean region does not support the notion of a progressive variation in the _CHIT1_ allele frequencies, as suggested by Malaguarnera et al. (2003). It is unlikely
that the frequencies found in some Mediterranean populations are linked to the disappearance of malaria. This parasitic disease has recently been eradicated in some areas, little more than
two generations ago (Hay et al. 2004), which is too limited a period of time to cause allele differences among the populations studied. These results seem to suggest that the H allele
originated among East Asian populations, where the highest frequencies are observed, and then spread to the West. The absence of this allele in sub-Saharan Africa seems to confirm this
hypothesis, but another scenario is possible. The mutant allele (if considered neutral with respect to natural selection) could have diffused out of Africa and been subsequently lost from
African populations due to genetic drift, while its frequency increased in Europe and Asia. However, for a definitive conclusion to be drawn, it would be necessary to clarify the roles of
the H allele and the selective pressures acting on _CHIT1_ or on flanking genes. The extent of linkage disequilibrium in this genic region should be investigated. The collection of data
about other Asiatic populations, for which there is an overwhelming lack of information from Turkey to India, may clarify the significance of the correlation between the allele frequencies
and longitude observed for all populations, and between Moran’s index and distance classes over 6,500 km. Moreover, a larger number of populations sampled would help to verify the validity
of the hypothesis that the H allele originated in Asia and the effects of natural selection or genetic drift, with a stronger spatial autocorrelation analysis. REFERENCES * Altarescu G,
Rudensky B, Abrahamov A, Goldfarb A, Rund D, Zimran A, Elstein D (2002) Plasma chitotriosidase activity in patients with beta-thalassemia. Am J Hematol 71:7–10 Article CAS PubMed Google
Scholar * Barbujani G (2000) Geographic patterns: how to identify them and why. Hum Biol 72:133–153 CAS PubMed Google Scholar * Barone R, Di Gregorio F, Romeo MA, Schilirò G, Pavone L
(1999) Plasma chitotriosidase activity in patients with β-thalassemia. Blood Cells Mol Dis 15:1–8 Article Google Scholar * Barone R, Bertrand G, Simpore J, Malaguarnera M, Musumeci S
(2001) Plasma chitotriosidase activity in beta-thalassemia major: a comparative study between Sicilian and Sardinian patients. Clin Chim Acta 306:91–96 Article CAS PubMed Google Scholar
* Barone R, Malaguarnera L, Angius A, Musumeci S (2002) Plasma chitotriosidase activity in patients with beta-thalassemia. Am J Hematol 71:7–10 Article CAS Google Scholar * Barone R,
Simpore J, Malaguarnera L, Pignatelli S, Musumeci S (2003) Plasma chitotriosidase activity in acute _Plasmodium falciparum_ malaria. Clin Chim Acta 331:79–85 Article CAS PubMed Google
Scholar * Boot RG, Renkema GH, Strijland A, van Zonneveld AJ, Aerts JM (1995) Cloning of cDNA encoding chitotriosidase, a human chitinase produced by macrophages. J Biol Chem
270:26252–26256 Article CAS PubMed Google Scholar * Boot RG, Renkema GH, Verhoek M, Strijland A, Bliek J, de Meulemeester TM, Mannens MM, Aerts JM (1998) The human chitotriosidase gene.
Nature of inherited enzyme deficiency. J Biol Chem 273:25680–25685 Article CAS PubMed Google Scholar * Boot RG, Blommaart EF, Swart E, Ghauharali-van der Vlugt K, Bijl N, Moe C, Place A,
Aerts JM (2001) Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J Biol Chem 276:6770–6778 Article CAS PubMed Google Scholar * Brown PJ (1981) New
considerations on the distribution of malaria, thalassemia, and glucose-6-phosphate dehydrogenase deficiency in Sardinia. Hum Biol 53:367–382 CAS PubMed Google Scholar * Cabrera-Salazar
MA, O’Rourke E, Henderson N, Wessel H, Barranger JA (2004) Correlation of surrogate markers of Gaucher disease. Implications for long-term follow up of enzyme replacement therapy. Clin Chim
Acta 344:101–107 Article CAS PubMed Google Scholar * Calafell F, Bertranpetit J (1994) Principal component analysis of gene frequencies and the origin of Basques. Am J Phys Anthropol
93:201–215 Article CAS PubMed Google Scholar * Canudas J, Cenarro A, Civeira F Garci-Otin AL, Aristegui R, Diaz C, Masramon X, Sol JM, Hernandez G, Pocovi M (2001) Chitotriosidase
genotype and serum activity in subjects with combined hyperlipidemia: effect of the lipid-lowering agents, atorvastatin and bezafibrate. Metabolism 50:447–450 Article CAS PubMed Google
Scholar * Chien YH, Chen JH, Hwu WL (2005) Plasma chitotriosidase activity and malaria. Clin Chim Acta 353:215 Article CAS PubMed Google Scholar * Choi EH, Zimmerman PA, Foster CB, Zhu
S, Kumaraswami V, Nutman TB, Chanock SJ (2001) Genetic polymorphisms in molecules of innate immunity and susceptibility to infection with _Wuchereria bancrofti_ in South India. Genes Immun
2:248–253 Article CAS PubMed Google Scholar * Choi EH, Taylor JG, Foster CB, Walsh TJ, Anttila VJ, Ruutu T, Palotie A, Chanock SJ (2005) Common polymorphisms in critical genes of innate
immunity do not contribute to the risk for chronic disseminated candidiasis in adult leukemia patients. Med Mycol 43:349–353 Article PubMed Google Scholar * Cliff AD, Ord JK (1973)
Spatial autocorrelation. Pion, London Google Scholar * Den Tandt WR, Van Hoof F (1996) Plasma methylumbelliferyl-tetra-_N_-acetyl-beta-d-chitotetraoside hydrolase as a parameter during
treatment of Gaucher patients. Biochem Mol Med 57:71–72 Article Google Scholar * Di Rosa M, Musumeci M, Scuto A, Musumeci S, Malaguarnera L (2005) Effect of interferon-gamma,
interleukin-10, lipopolysaccharide and tumor necrosis factor-alpha on chitotriosidase synthesis in human macrophages. Clin Chem Lab Med 43:499–502 Article CAS PubMed Google Scholar *
Ewens WJ (1972) The sampling theory of selectively neutral alleles. Theor Popul Biol 3:82–112 Google Scholar * Fermi C (1934) Regioni malariche. Decadenza, risanamento e spesa “Sardegna”,
vol 1. Tipografia Editrice di Roma SA, Roma * Fermi C (1938) Provincia di Nuoro. Malaria, danni economici, risanamento e proposte per il suo risorgimento, vol 2. Gallizzi, Sassari *
Gianfrancesco F, Musumeci S (2004) The evolutionary conservation of the human chitotriosidase gene in rodents and primates. Cytogenet Genome Res 105:54–56 Article CAS PubMed Google
Scholar * Grosso S, Margollicci MA, Bargagli E, Buccoliero QR, Perrone A, Galimberti D, Morgese G, Balestri P, Rottoli P (2004) Serum levels of chitotriosidase as a marker of disease
activity and clinical stage in sarcoidosis. Scand J Clin Lab Invest 64:57–62 Article CAS PubMed Google Scholar * Guo Y, He W, Boer AM, Wevers RA, de Bruijn AM, Groener JE, Hollak CE,
Aerts JM, Galjaard H, van Diggelen OP (1995) Elevated plasma chitotriosidase activity in various lysosomal storage disorders. J Inherit Metab Dis 18:717–722 Article CAS PubMed Google
Scholar * Hay SI, Guerra CA, Tatem AJ, Noor AM, Snow RW (2004) The global distribution and population at risk of malaria: past, present, and future. Lancet Infect Dis 4:327–336 Article
PubMed PubMed Central Google Scholar * Hise AG, Hazlett FE, Bockarie MJ, Zimmerman PA, Tisch DJ, Kazura JW (2003) Polymorphisms of innate immunity genes and susceptibility to lymphatic
filariasis. Genes Immun 4:524–527 Article CAS PubMed Google Scholar * Hollak CE, van Weely S, van Oers MHJ, Aerts JMPG (1994) Marked elevation of plasma chitotriosidase activity. A novel
hallmark in Gaucher disease. J Clin Invest 93:1288–1292 Article CAS PubMed PubMed Central Google Scholar * Lancaster A, Nelson PM, Single RM, Meyer D, Thomson G (2003) Pypop: a
software framework for population genomics: analyzing large-scale multi-locus genotype data. In: Altman RB et al (eds) Pacific symposium on biocomputing eight. World Scientific, Singapore,
pp 514–525 * Lin SY, Tzeng SH, Chang YM, Siauw CP, Chen PH (1991) Imported case of malaria in Taiwan: analysis of 11 cases. J Formos Med Assoc 90:308–311 CAS PubMed Google Scholar * Logan
JA (1957) Il progetto Sardegna: un esperimento di eradicazione del vettore indigeno della malaria, Edizione Italiana. The Johns Hopkins, Baltimore Google Scholar * Malaguarnera L, Simporè
J, Prodi DA, Angius A, Sassu A, Persico I, Barone R, Musumeci S (2003) A 24-bp duplication in exon ten of human chitotriosidase gene from the sub-Saharan to the Mediterranean area: role of
parasitic diseases and environmental conditions. Genes Immun 4:570–574 Article CAS PubMed Google Scholar * Malaguarnera L, Musumeci M, Licata F, Di Rosa M, Messina A, Musumeci S (2004)
Prolactin induces chitotriosidase gene expression in human monocyte-derived macrophages. Immunol Lett 94:57–63 Article CAS PubMed Google Scholar * Piras IS, Melis A, Ghiani ME, Calò CM,
Vona G (2006) Variabilità del gene _CHIT_ nel Mediterraneo, I SIBE Congress, Florence (Italy), 4–7/09/2006. (http://www.dbs.unica.it/temp/poster_sito_internet.ppt) * Raymond M, Rousset F
(1995) Genepop 1.2: population genetics software for exact tests and ecumenicim. J Hered 86:248–249 Article Google Scholar * Renkema GH, Boot RG, Strjland A, Donker-Kooplam WE, van der
Berg M, Muijers AO, Aerts JM (1997) Synthesis, sorting and processing into distinct isoforms of human macrophage chitotriosidase. Eur J Biochem 244:279–285 Article CAS PubMed Google
Scholar * Rodrigues MR, Sa Miranda MC, Amaral O (2004) Allelic frequency determination of the 24-bp chitotriosidase duplication in the Portuguese population by real-time PCR. Blood Cells
Mol Dis 33:362–364 Article CAS PubMed Google Scholar * Sanna E, Cosseddu GG, Floris G, Liguori A, Silvetti M (1997) Micromapping the distribution of G6PD deficiency in Sardinia with data
collected from the 1950s to the 1980s. In: Green LS (ed) Adaptation to malaria. The interaction of biology and culture. Gordon and Breach, New York, pp 293–305 * Siniscalco M, Bernini L,
Filippi G, Latte B, Meera Khan P, Piomelli S, Rattazzi M (1966). Population genetics of haemoglobin variants, thalassaemia and glucose-6-phosphate dehydrogenase deficiency, with particular
reference to the malaria hypothesis. Bull World Health Organ 34:379–393 CAS PubMed PubMed Central Google Scholar * Slatkin M (1994) An exact test for neutrality based on the Ewens
sampling distribution. Genet Res 64:71–74 Article CAS PubMed Google Scholar * Sokal RR, Oden NL (1978) Spatial autocorrelation in biology. 1. Methodology. Biol J Linn Soc 10:199–249
Article Google Scholar * Sokal RR, Harding RM, Oden NL (1989) Spatial patterns of human gene frequencies in Europe. Am J Phys Anthropol 80:267–294 Article CAS PubMed Google Scholar *
van Eijk M, van Roomen CP, Renkema GH, Bussink AP, Andrews L, Blommaart EF, Sugar A, Verhoeven AJ, Boot RG, Aerts JM (2005) Characterization of human phagocyte-derived chitotriosidase, a
component of innate immunity. Int Immunol 17:1505–1512 Article PubMed Google Scholar * Varesi L, Memmi M, Cristofari MC, Mameli GE, Calo CM, Vona G (2000) Mitochondrial control-region
sequence variation in the Corsican population, France. Am J Hum Biol 12:339–351 Article PubMed Google Scholar * Vona G (1997) The peopling of Sardinia (Italy): history and effects. Int J
Anthropol 12:71–87 Article Google Scholar * Wartenberg D (1989) SAAP—Spatial Autocorrelation Analysis Program. Rutgers University, Piscataway, NJ * Watterson G (1978) The homozygosity test
of neutrality. Genetics 88:405–417 Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This research was supported by grants from the University of
Cagliari; 60% (G.V.) AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Experimental Biology, University of Cagliari, SS 554, km 4,500, 09042, Monserrato (CA), Italy Ignazio Piras,
Alessandra Melis, Maria Elena Ghiani, Carla Maria Calò & Giuseppe Vona * Department of Human Genetics, University of Corsica, Corte, France Alessandra Falchi & Laurent Varesi *
Department of Experimental Evolutionistic Biology, University of Bologna, Bologna, Italy Donata Luiselli * Department of Animal Biology, University of Barcelona, Barcelona, Spain Pedro Moral
Authors * Ignazio Piras View author publications You can also search for this author inPubMed Google Scholar * Alessandra Melis View author publications You can also search for this author
inPubMed Google Scholar * Maria Elena Ghiani View author publications You can also search for this author inPubMed Google Scholar * Alessandra Falchi View author publications You can also
search for this author inPubMed Google Scholar * Donata Luiselli View author publications You can also search for this author inPubMed Google Scholar * Pedro Moral View author publications
You can also search for this author inPubMed Google Scholar * Laurent Varesi View author publications You can also search for this author inPubMed Google Scholar * Carla Maria Calò View
author publications You can also search for this author inPubMed Google Scholar * Giuseppe Vona View author publications You can also search for this author inPubMed Google Scholar
CORRESPONDING AUTHOR Correspondence to Ignazio Piras. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Piras, I., Melis, A., Ghiani, M.E. _et al._ Human
_CHIT1_ gene distribution: new data from Mediterranean and European populations. _J Hum Genet_ 52, 110 (2007). https://doi.org/10.1007/s10038-006-0086-1 Download citation * Received: 21 July
2006 * Accepted: 24 October 2006 * Published: 15 November 2006 * DOI: https://doi.org/10.1007/s10038-006-0086-1 SHARE THIS ARTICLE Anyone you share the following link with will be able to
read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative KEYWORDS * _CHIT1_ * H allele * Spatial autocorrelation * Malaria * Mediterranean populations