Risk alleles for chronic hepatitis b are associated with decreased mrna expression of hla-dpa1 and hla-dpb1 in normal human liver
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT A genome-wide association study identified single nucleotide polymorphisms (SNPs) rs3077 and rs9277535 located in the 3′ untranslated regions of human leukocyte antigen (HLA) class
II genes _HLA-DPA1_ and _HLA-DPB1_, respectively, as the independent variants most strongly associated with chronic hepatitis B. We examined whether these SNPs are associated with mRNA
expression of _HLA-DPA1_ and _HLA-DPB1_. We identified gene expression-associated SNPs (eSNPs) in normal liver samples obtained from 651 individuals of European ancestry by integrating
genotype (∼650 000 SNPs) and gene expression (>39 000 transcripts) data from each sample. We used the Kruskal–Wallis test to determine associations between gene expression and genotype.
To confirm findings, we measured allelic expression imbalance (AEI) of complementary DNA compared with DNA in liver specimens from subjects who were heterozygous for rs3077 and rs9277535. On
a genome-wide basis, rs3077 was the SNP most strongly associated with _HLA-DPA1_ expression (_p_=10−48), and rs9277535 was strongly associated with _HLA-DPB1_ expression (_p_=10−15).
Consistent with these gene expression associations, we observed AEI for both rs3077 (_p_=3.0 × 10−7; 17 samples) and rs9277535 (_p_=0.001; 17 samples). We conclude that the variants
previously associated with chronic hepatitis B are also strongly associated with mRNA expression of _HLA-DPA1_ and _HLA-DPB1_, suggesting that expression of these genes is important in
control of HBV. SIMILAR CONTENT BEING VIEWED BY OTHERS POLYMORPHISMS WITHIN _DIO2_ AND _GADD45A_ GENES INCREASE THE RISK OF LIVER DISEASE PROGRESSION IN CHRONIC HEPATITIS B CARRIERS Article
Open access 14 April 2023 INTEGRATIVE COMMON AND RARE VARIANT ANALYSES PROVIDE INSIGHTS INTO THE GENETIC ARCHITECTURE OF LIVER CIRRHOSIS Article Open access 17 April 2024 GENOME-WIDE COPY
NUMBER VARIATION ANALYSIS OF HEPATITIS B INFECTION IN A JAPANESE POPULATION Article Open access 08 June 2021 INTRODUCTION Chronic infection with hepatitis B virus (HBV) is the most common
cause of liver cancer worldwide, as well as a major risk factor for development of cirrhosis and end-stage liver disease.1 The hepatitis B vaccine is highly effective in preventing new
infections, but ∼360 million people still suffer from chronic hepatitis B and there are ∼600 000 annual deaths from HBV-related causes.1 Recent genome-wide association studies identified
single nucleotide polymorphisms (SNPs) located within the human leukocyte antigen (HLA) class II genes _HLA-DPA1_ and _HLA-DPB1_ to be associated with chronic hepatitis B.2, 3 Replication
studies performed on two of these SNPs (rs3077 and rs9277535) confirmed and strengthened results from the genome-wide association studies.2 Class II HLA genes encode proteins expressed on
the surface of antigen-presenting cells such as macrophages, dendritic cells and B cells, and thereby have a critical role in presentation of antigens to CD4+ T-helper lymphocytes. HLA genes
have many structural variants that have been linked to immune response to infectious agents,4 but genetic variants that influence HLA mRNA expression might also affect antigen presentation
and many ‘gene expression-associated SNPs’ (eSNPs) have been found for HLA genes.5, 6 An integrated approach combining genotype information with genome-wide gene expression data in relevant
tissues can identify genetic variations that are both regulatory and disease causing.6, 7 In this study, we examined whether SNPs implicated in chronic hepatitis B are associated with mRNA
expression in liver samples from the Human Liver Cohort, a large study of genotype and gene expression in normal liver.6 Additionally, we performed confirmatory allelic expression imbalance
(AEI) studies in human liver and peripheral blood. We found that SNPs associated with chronic hepatitis B are strongly associated with decreased expression of important antigen-presenting
molecules, _HLA-DPA1_ and _HLA-DPB1_. RESULTS GENETIC VARIANTS AND MRNA EXPRESSION A published genome-wide association study for chronic hepatitis B identified 11 disease-associated SNPs;
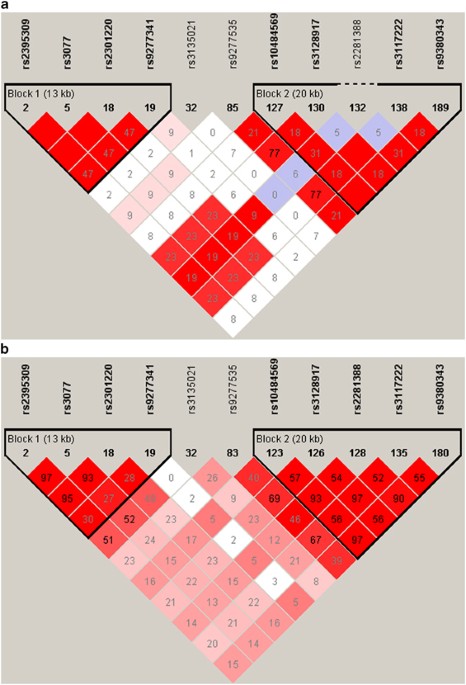
confirmatory replication studies were performed on rs3077 and rs9277535 (ref. 2). SNP rs3077 is located in the 3′ untranslated region (3′ UTR) of _HLA-DPA1,_ rs2395309 lies ∼6.5 kb
downstream of _HLA-DPA1_ and rs2301220 is found in the first intron of _HLA-DPA1._ These SNPs are in complete linkage disequilibrium (LD) in Europeans (CEU HapMap samples, _r__2_=1.0; Figure
1a) and in near complete LD in Asians (JPT+CHB HapMap samples, _r__2_=0.93–0.97, Figure 1b). Among the ∼650 000 SNPs that were genotyped and examined for association with expression of
_HLA-DPA1_, rs2395309, rs3077 and rs2301220 were the variants most strongly associated with mRNA expression of this gene (_p_∼10−48; Table 1). Of the eight remaining SNPs that were
associated with chronic hepatitis B,2 five were associated with expression of _HLA-DPA1_ as well (Table 1). Figure 2a shows odds ratios for chronic hepatitis B2 and differences in mean-log
gene expression by rs3077 genotypes. Gene expression was measured relative to a pool of control liver samples,6 and the risk genotype for chronic HBV (GG) is set to zero as a reference. The
rs3077-G allele was associated with both higher risk of chronic hepatitis B and lower expression of _HLA-DPA1_ in a pattern consistent with an additive genetic model. Similarly, SNP
rs9277535, located within the 3′ UTR of _HLA-DPB1_ (∼22 kb from rs3077), was strongly associated with both increased risk of chronic hepatitis B and decreased expression of _HLA-DPB1_
(_p_=10−15; Table 1, Figure 2b). There is only weak LD between rs9277535 and rs3077 in Europeans (_r__2_=0.09, D′=0.50; HapMap CEU; Figure 1a) and Asians (_r__2_=0.24, D′=0.54; HapMap
JPT+CHB; Figure 1b), suggesting that effects of these SNPs are likely to be independent. To confirm that rs3077 and rs9277535 are associated with increased mRNA expression of their
respective genes, we also examined genotype and gene expression data from a publically available database of 400 Epstein-Barr virus-transformed lymphoblastoid cell lines obtained from
British children with asthma.5 Among these subjects, rs3077 was strongly associated with _HLA-DPA1_ expression (_p_=10−21) and rs9277535 with _HLA-DPB1_ expression (_p_=10−12). ALLELIC
EXPRESSION IMBALANCE In the Human Liver Cohort, mRNA expression was measured by a microarray. It has been suggested that genetic and structural variations within the HLA region could affect
efficiency of hybridization of microarray probes,8, 9 and, if so, differences in mRNA levels could reflect differences in detection efficiency rather than true differences in gene
expression. We reasoned that if a genetic variation is truly associated with altered gene expression in _cis_, samples from heterozygous individuals should display AEI in complementary DNA
(cDNA) when compared with DNA.10 The location of rs3077 and rs9277535 in transcribed (3′ UTR) regions of _HLA-DPA1_ and _HLA-DPB1_ makes these SNPs good candidates for AEI testing.
Furthermore, AEI Taqman assays for these SNPs were designed to avoid any other underlying genetic variation in the amplicons. Therefore, AEI is an independent test for possible differences
in gene expression associated with genetic variants. Consistent with the mRNA expression associations found in the Human Liver Cohort, we observed AEI for both rs3077 (liver tissue (_n_=17),
_p_=3.0 × 10−7, Figure 3a) and monocytes ((_n_=22), _p_=2.0 × 10−8, Figure 3b) and rs9277535 (liver tissue (_n_=17), _p_=0.001, Figure 3c) and monocytes ((_n_=17), _p_=0.04, Figure 3d). For
both SNPs, the proportion of non-risk allele A was significantly increased in cDNA compared with DNA of heterozygous samples, indicating reduced expression of the risk alleles. AEI for
rs3077 was not explained by rs9277535 and vice versa (Supplementary Figure 1), although the statistical power for those comparisons was more limited than for the primary comparisons.
Therefore, it is likely that rs3077 and rs9277535 (or variants in high LD with these SNPs) exhibit independent effects on the expression of _HLA-DPA1_ and _HLA-DPB1_, respectively.
DISCUSSION On the basis of findings for both genome-wide mRNA expression and allelic expression imbalance, it seems that SNPs rs3077 and rs9277535 are strongly associated with regulation of
_HLA-DPA1_ and _HLA-DPB1_, respectively. Previous studies found that these SNPs were highly associated with chronic HBV infection.2, 3 Together, these independent studies strongly implicate
lower expression of _HLA-DPA1_ and _HLA-DPB1_ with increased risk of chronic HBV. HLA-DPA1 and HLA-DPB1 are expressed on the surface of antigen-presenting cells. In the liver, expression of
these proteins is likely to be limited to a small population of Kupffer cells, the resident macrophages of the liver. Kupffer cells are derived from blood monocytes, and we observed evidence
for AEI in both monocytes and liver samples. We found that mRNA expression of both _HLA-DPA1_ and _HLA-DPB1_ was low in total human liver tissue, suggesting that it might be limited to a
specific sub-population of cells. We attempted to quantify protein expression in lysates from total liver tissue, but failed to detect a measurable signal (data not shown). Similarly,
HLA-DPA1 and HLA-DPB1 protein expression was undetectable in liver tissue examined by antibody-based proteomic methods.11 It is intriguing that the genetic risk variants associated with
chronic hepatitis B affect expression of the alpha (DPA) and beta (DPB) chains of the same antigen-presenting complex, as this suggests that insufficient expression of either or both of
these chains might result in the same phenotype. Kamatani _et al._2 reported that chronic hepatitis B was associated with haplotypes comprised of rs3077-G, rs9277535-G and certain structural
variants of _HLA-DPA1_ and _HLA-DPB1._ Our findings do not exclude a role for structural variants of _HLA-DPA1_ and _HLA-DPB1_ in chronic hepatitis B, as viral control of HBV could be
affected by both regulatory and structural variants. For example, functional variants of _MBL2_, which has an important role in host defense against HIV-1 and other infectious agents,
include both regulatory and structural variants.12 On a genome-wide basis, rs3077 is the variant most strongly associated with risk of chronic hepatitis B and the variant most strongly
associated with expression of _HLA-DPA1._ It seems highly likely, therefore, that expression of _HLA-DPA1_ has a role in the clearance of HBV. Similarly, the association of rs9277535 with
chronic HBV and with _HLA-DPB1_ expression suggests that _HLA-DPB1_ expression also contributes to chronic HBV infection. Kamatani _et al._2 noted that the HBV risk alleles for rs3077 and
rs9277535 are more common in Asians than Europeans (fitting the global pattern for the epidemiology of chronic hepatitis B). LD patterns in this genomic region are very similar for
populations of Asian or European ancestry groups (Figures 1a and b), indicating that the relationships between SNPs in this region are comparable between these two ancestral groups even
though the allele frequencies differ. We could not determine why the rs3077 and rs9277535 variants are associated with decreased mRNA expression of _HLA-DPA1_ and _HLA-DPB1_ in liver, but
variants within the 3′ UTR may affect mRNA stability through binding of regulatory factors or regulation by microRNAs.13 SNP rs3077 was found to be associated with methylation level of
_HLA-DPB1_ and _HLA-DPA1_ in adult cerebellum samples studied by Zhang _et al._14 (see Supplementary Table 5 of that paper). It may also be noteworthy that rs9277535 was linked to two
different copy-number variation regions in European HapMap subjects.15 We did not observe any obvious factors that could explain differential expression of _HLA-DPA1_ and _HLA-DPB1_ mRNA,
but future studies should address this question. SNPs rs3077and rs9277535 have been associated with other diseases. The rs9277535 variant has been linked to primary biliary cirrhosis.16 Both
rs3077 and rs9277535 have been associated with rheumatoid arthritis,17 although other SNPs in the major histocompatibility complex class II region were more strongly associated with that
disease. In conclusion, genetic variants previously associated with chronic hepatitis B2 are also associated with decreased expression of an antigen-presenting complex that consists of
_HLA-DPA1_ and _HLA-DPB1_ chains. Together, these independent studies strongly implicate lower expression of _HLA-DPA1_ and _HLA-DPB1_ as a factor for increased risk of chronic hepatitis B.
Other recent studies have demonstrated a relationship between expression of _HLA-C_ and control of HIV.18, 19 It is possible, therefore, that HLA expression has a role in control of a range
of viruses. More broadly, our findings lend support to the concept that SNPs identified through an integrated genomic approach can provide both confirmation and functional insights for
disease associations.6, 20 If greater expression of _HLA-DPA1_ and _HLA-DPB1_ facilitate clearance of HBV infection, development of therapies that increase expression of these genes might
aid in treatment of chronic hepatitis B. METHODS GENOME-WIDE ASSOCIATION OF SNPS WITH MRNA EXPRESSION As previously described,6 the Human Liver Cohort identifies eSNPs by integrating
genotype and gene expression data for liver samples without evidence of liver disease. The current analysis is based on 651 liver samples obtained from patients of non-Hispanic European
ancestry. Genome-wide gene expression was measured with custom-designed Agilent (Agilent Technologies, Inc., Santa Clara, CA, USA) arrays (>39 000 transcripts), and genotyping was
performed with the Illumina (San Diego, CA, USA) Sentrix HumanHap650Y genotyping chips.6 The Kruskal–Wallis test was used to determine association between adjusted expression traits
according to an additive genetic model. False discovery rate threshold was set at 10%. Associations were considered significant at _p_<5.0 × 10−5 for _cis_–factors (probe to SNP distance
<1 Mb) and _p_<1.0 × 10−8 for _trans_ –factors.6 Only SNPs used both in genome-wide association studies for chronic HBV2 and the mRNA expression study6 were used in this analysis. For
each of the SNPs, gene expression values were compared with the referent non-risk genotypes. ALLELIC EXPRESSION IMBALANCE Liver tissue samples for AEI studies were non-cancerous specimens
provided by the Liver Tissue Cell Distribution System (LTCDS). Anonymized fresh peripheral blood samples from healthy donors were provided by the Blood Bank at the NIH. Monocytes were
purified from fresh blood with CD14+-coated magnetic MicroBeads with AutoMacs (Miltenyi Biotec, Auburn, CA, USA) as previously described.21 DNA from all samples was prepared with DNAeasy kit
(Qiagen, Valencia, CA, USA) and RNA was prepared with Trizol reagent (Invitrogen, Carlsbad, CA, USA) followed by RNAeasy kit (Qiagen). The AEI was quantified with the allelic discrimination
genotyping assays C__11916951_10 for rs3077 and a custom-designed assay for rs9277535. To ensure that the signal was specific for cDNA and was not derived from residual DNA in these
samples, a ‘no reverse-transcriptase’ control reaction was tested initially. Genotyping for all DNA and cDNA samples was performed in duplicate on the 7900 Sequence Detection System (ABI,
Foster City, CA, USA). AEI experiments were carried out as previously described.22, 23 A standard curve was prepared based on 10 dilutions of two homozygous DNA samples prepared from liver
tissue, representing 5, 15, 25, 35, 45, 55, 65, 75, 85 and 95% of allele ‘X’, based on the standard curve. The potential AEI in samples heterozygous for rs3077 and rs9277535 was evaluated by
comparing the proportion of allele ‘X’ in DNA with that in cDNA with a paired two-sided _T_-test. To evaluate the potential effect of one SNP on AEI for the other (for example, effect of
rs3077 on AEI for rs9277535), we tested whether the proportion of allele ‘X’ in samples heterozygous for rs9277535 differed by the genotype of rs3077. Using an F-test, we compared the
proportion of allele ‘A’ in all samples that were heterozygous for rs9277535 with the proportion of allele ‘A’ in samples that were heterozygous for rs9277535, but homozygous for rs3077.
REFERENCES * Shepard CW, Simard EP, Finelli L, Fiore AE, Bell BP . Hepatitis B virus infection: epidemiology and vaccination. _Epidemiol Rev_ 2006; 28: 112–125. Article PubMed Google
Scholar * Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N _et al_. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic
hepatitis B in Asians. _Nat Genet_ 2009; 41: 591–595. Article CAS PubMed Google Scholar * Matsuura K, Tanaka Y, Nishida N, Hige S, Asahina Y, Ito K _et al_. Genome-wide association
study identifies genetic variants in the HLA-DP locus associated with chronic hepatitis B. _J Hepatol_ 2010; 52 (Supplement 1): S282–S283. Article Google Scholar * MHC sequencing
consortium. Complete sequence and gene map of a human major histocompatibility complex. _Nature_ 1999; 401: 921–923. Article Google Scholar * Dixon AL, Liang L, Moffatt MF, Chen W, Heath
S, Wong KC _et al_. A genome-wide association study of global gene expression. _Nat Genet_ 2007; 39: 1202–1207. Article CAS PubMed Google Scholar * Schadt EE, Molony C, Chudin E, Hao K,
Yang X, Lum PY _et al_. Mapping the genetic architecture of gene expression in human liver. _PLoS Biol_ 2008; 6: e107. Article PubMed PubMed Central Google Scholar * Emilsson V,
Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J _et al_. Genetics of gene expression and its effect on disease. _Nature_ 2008; 452: 423–428. Article CAS PubMed Google Scholar *
Alberts R, Terpstra P, Li Y, Breitling R, Nap J-P, Jansen RC . Sequence polymorphisms cause many false cis eQTLs. _PLoS ONE_ 2007; 2: e622. Article PubMed PubMed Central Google Scholar *
Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M . Mapping complex disease traits with global gene expression. _Nat Rev Genet_ 2009; 10: 184–194. Article CAS PubMed PubMed Central
Google Scholar * Johnson AD, Zhang Y, Papp AC, Pinsonneault JK, Lim JE, Saffen D _et al_. Polymorphisms affecting gene transcription and mRNA processing in pharmacogenetic candidate genes:
detection through allelic expression imbalance in human target tissues. _Pharmacogenet Genomics_ 2008; 18: 781–791. Article CAS PubMed PubMed Central Google Scholar * Uhlen M, Bjorling
E, Agaton C, Szigyarto CA, Amini B, Andersen E _et al_. A human protein atlas for normal and cancer tissues based on antibody proteomics. _Mol Cell Proteomics_ 2005; 4: 1920–1932. Article
CAS PubMed Google Scholar * Garred P, Larsen F, Seyfarth J, Fujita R, Madsen HO . Mannose-binding lectin and its genetic variants. _Genes Immun_ 2006; 7: 85–94. Article CAS PubMed
Google Scholar * Prokunina L, Alarcon-Riquelme ME . Regulatory SNPs in complex diseases: their identification and functional validation. _Expert Rev Mol Med_ 2004; 6: 1–15. Article PubMed
Google Scholar * Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W _et al_. Genetic control of individual differences in gene-specific methylation in human brain. _Am J Hum Genet_ 2010;
86: 411–419. Article CAS PubMed PubMed Central Google Scholar * Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y _et al_. Origins and functional impact of copy number variation
in the human genome. _Nature_ 2010; 464: 704–712. Article CAS PubMed Google Scholar * Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y _et al_. Primary biliary cirrhosis associated with
HLA, IL12A, and IL12RB2 Variants. _N Engl J Med_ 2009; 360: 2544–2555. Article CAS PubMed PubMed Central Google Scholar * Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B
_et al_. TRAF1-C5 as a risk locus for rheumatoid arthritis—a genomewide study. _N Engl J Med_ 2007; 357: 1199–1209. Article CAS PubMed PubMed Central Google Scholar * Thomas R, Apps R,
Qi Y, Gao X, Male V, O’HUigin C _et al_. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. _Nat Genet_ 2009; 41: 1290–1294. Article CAS
PubMed PubMed Central Google Scholar * Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M _et al_. A whole-genome association study of major determinants for host control of
HIV-1. _Science_ 2007; 317: 944–947. Article CAS PubMed PubMed Central Google Scholar * Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R _et al_. Genome-wide
associations of gene expression variation in humans. _PLoS Genet_ 2005; 1: e78. Article PubMed PubMed Central Google Scholar * Prokunina-Olsson L, Welch C, Hansson O, Adhikari N, Scott
L, Usher N _et al_. Tissue-specific alternative splicing of TCF7L2. _Hum Mol Genet_ 2009; 18: 3795–3804. Article CAS PubMed PubMed Central Google Scholar * Cheung VG, Spielman RS, Ewens
KG, Weber TM, Morley M, Burdick JT . Mapping determinants of human gene expression by regional and genome-wide association. _Nature_ 2005; 437: 1365–1369. Article CAS PubMed PubMed
Central Google Scholar * Dennis MY, Paracchini S, Scerri TS, Prokunina-Olsson L, Knight JC, Wade-Martins R _et al_. A common variant associated with dyslexia reduces expression of the
KIAA0319 gene. _PLoS Genet_ 2009; 5: e1000436. Article PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We acknowledge the assistance of Sabrina Chen
(Information Management Services, Silver Spring, Maryland) and Myhanh Dotrang (CSC, Rockville, Maryland) with databases, David Check with graphics and Natalia Orduz with the AEI studies.
Liver tissue samples for AEI testing were kindly provided by the Liver Tissue Cell Distribution System under NIH Contract #N01-DK-7-0004/HHSN267200700004C. This research was supported by the
Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics. It was funded in part with federal funds from the
National Cancer Institute, National Institutes of Health, under contract Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of
the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. AUTHOR INFORMATION AUTHORS AND
AFFILIATIONS * Department of Health and Human Services, Infections and Immunoepidemiology Branch, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA T R O'Brien
* Department of Health and Human Services, Laboratory of Translational Genomics, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA I Kohaar & L
Prokunina-Olsson * Department of Health and Human Services, Biostatistics Branch, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA R M Pfeiffer * Division of
Cancer Epidemiology and Genetics, Department of Health and Human Services, Human Genetics Program, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA D Maeder &
M Yeager * NCI Core Genotyping Facility, SAIC-Frederick, Inc., NCI-Frederick, Gaithersburg, MD, USA M Yeager * Sage Bionetworks, Seattle, WA, USA E E Schadt * Pacific Biosciences, Menlo
Park, CA, USA E E Schadt Authors * T R O'Brien View author publications You can also search for this author inPubMed Google Scholar * I Kohaar View author publications You can also
search for this author inPubMed Google Scholar * R M Pfeiffer View author publications You can also search for this author inPubMed Google Scholar * D Maeder View author publications You can
also search for this author inPubMed Google Scholar * M Yeager View author publications You can also search for this author inPubMed Google Scholar * E E Schadt View author publications You
can also search for this author inPubMed Google Scholar * L Prokunina-Olsson View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR
Correspondence to T R O'Brien. ETHICS DECLARATIONS COMPETING INTERESTS Eric E Schadt is Chief Scientific Officer of Pacific Biosciences. ADDITIONAL INFORMATION Supplementary Information
accompanies the paper on Genes and Immunity website SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (DOC 75 KB) RIGHTS AND PERMISSIONS This work is licensed under the Creative Commons
Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/ Reprints and permissions ABOUT
THIS ARTICLE CITE THIS ARTICLE O'Brien, T., Kohaar, I., Pfeiffer, R. _et al._ Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of _HLA-DPA1_ and
_HLA-DPB1_ in normal human liver. _Genes Immun_ 12, 428–433 (2011). https://doi.org/10.1038/gene.2011.11 Download citation * Received: 24 August 2010 * Revised: 11 November 2010 * Accepted:
11 November 2010 * Published: 24 February 2011 * Issue Date: September 2011 * DOI: https://doi.org/10.1038/gene.2011.11 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative KEYWORDS * chronic hepatitis B * HLA * gene expression * genetics * genomics