Pocket the difference | Nature Reviews Molecular Cell Biology
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Access through your institution Buy or subscribe Kathryn Ferguson and colleagues at the University of Pennsylvania, and Susan Lietzke and co-workers up the coast at the University of
Massachusetts, both chose the PH domain from the PtdIns(3,4,5)P3-specific protein Grp1 (also known as ARNO3 or cytohesin 3) as the subject of their crystallographic analysis. Ferguson _et
al_. also solved the structure of DAPP1 (PHISH), which binds PtdIns(3,4,5)P3 and PtdIns(3,4)P2 with similar affinity. Lietzke and colleagues compared features of the Grp1 PH domain with the
promiscuous PH domain from protein kinase B (AKT). In both studies, inositol-1,3,4,5-tetrakisphosphate (Ins(1,3,4,5)P4) — the water-soluble headgroup of PtdIns(3,4,5)P3 — was used to
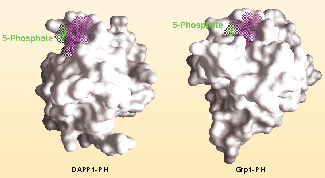
crystallize the ligand-bound PH domains. It was no surprise that the PH domains of both Grp1 and DAPP1 are β-sandwiches, just like previously solved PH domains, but Grp1 has an extra two
β-strands. These form a snug pocket accommodating the 5-phosphate (see picture), which simply isn't there in the non-discriminating PH domains of DAPP1 and protein kinase B. This
difference explains how Grp1 binds PtdIns(3,4,5)P3, but how does it stop PtdIns(3,4)P2 from binding? The answer lies in the number of hydrogen bonds necessary to stabilize the interaction.
Ferguson and colleagues find that both Grp1 and DAPP1 make a total of 14 side-chain hydrogen bonds with Ins(1,3,4,5)P4, but in Grp1 these are equally distributed among the 3-, 4- and
5-phosphates, whereas in DAPP1 there are no hydrogen bonds to the 5-phosphate. So, when PtdIns(3,4)P2 enters Grp1's PH domain, there's nothing for this extra pocket to grab hold
of, and Grp1 can't hang on tightly enough to just the 3- and 4-phosphates. This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your
institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access
to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read
our FAQs * Contact customer support ORIGINAL RESEARCH PAPERS * Ferguson, K. M. et al. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. _ Mol. Cell_
6, 373–384 ( 2000) Article CAS Google Scholar * Lietzke, S. E. _ et al_. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. _Mol. Cell_ 6, 385– 394 (2000)
Article CAS Google Scholar * REVIEWS Lemmon, M. A. & Ferguson, K. M. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. _ Biochem. J._ 350, 1–18 ( 2000) Article
CAS Google Scholar * Chan, T. O. _ et al_. AKT/PKB and other D3 phosphoinositide-regulated kinases: kinase activation by phosphoinositide-dependent phosphorylation. _Annu. Rev. Biochem._
68, 965–1014 (1999) Article CAS Google Scholar Download references Authors * Cath Brooksbank View author publications You can also search for this author inPubMed Google Scholar RIGHTS
AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Brooksbank, C. Pocket the difference. _Nat Rev Mol Cell Biol_ 1, 9 (2000). https://doi.org/10.1038/35036032
Download citation * Issue Date: 01 October 2000 * DOI: https://doi.org/10.1038/35036032 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative