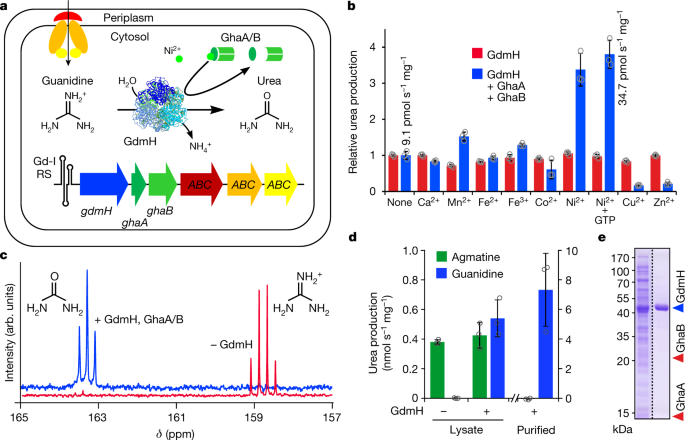
Discovery of a ni2+-dependent guanidine hydrolase in bacteria
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Nitrogen availability is a growth-limiting factor in many habitats1, and the global nitrogen cycle involves prokaryotes and eukaryotes competing for this precious resource. Only
some bacteria and archaea can fix elementary nitrogen; all other organisms depend on the assimilation of mineral or organic nitrogen. The nitrogen-rich compound guanidine occurs widely in
nature2,3,4, but its utilization is impeded by pronounced resonance stabilization5, and enzymes catalysing hydrolysis of free guanidine have not been identified. Here we describe the
arginase family protein GdmH (Sll1077) from _Synechocystis_ sp. PCC 6803 as a Ni2+-dependent guanidine hydrolase. GdmH is highly specific for free guanidine. Its activity depends on two
accessory proteins that load Ni2+ instead of the typical Mn2+ ions into the active site. Crystal structures of GdmH show coordination of the dinuclear metal cluster in a geometry typical for
arginase family enzymes and allow modelling of the bound substrate. A unique amino-terminal extension and a tryptophan residue narrow the substrate-binding pocket and identify homologous
proteins in further cyanobacteria, several other bacterial taxa and heterokont algae as probable guanidine hydrolases. This broad distribution suggests notable ecological relevance of
guanidine hydrolysis in aquatic habitats. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access
through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to
this journal Receive 51 print issues and online access $199.00 per year only $3.90 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy
now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS MARINE PICOCYANOBACTERIAL PHND1 SHOWS SPECIFICITY FOR VARIOUS PHOSPHORUS SOURCES BUT LIKELY REPRESENTS A CONSTITUTIVE INORGANIC PHOSPHATE
TRANSPORTER Article Open access 22 April 2023 ASSIMILATORY SULFATE REDUCTION IN THE MARINE METHANOGEN _METHANOTHERMOCOCCUS THERMOLITHOTROPHICUS_ Article Open access 05 June 2023
Α-CYANOBACTERIA POSSESSING FORM IA RUBISCO GLOBALLY DOMINATE AQUATIC HABITATS Article Open access 18 July 2022 DATA AVAILABILITY Structure coordinates of GdmH from _Synechocystis_ sp.
PCC6803 and experimental structure factor amplitudes have been deposited with the Protein Data Bank as entries 7OI1 and 7ESR, for space groups _C_2 and _R_32, respectively. X-ray diffraction
images have been deposited with Zenodo, at which https://doi.org/10.5281/zenodo.4750963 corresponds to PDB entry 7OI1 and https://doi.org/10.5281/zenodo.4750940corresponds to 7ESR. The raw
data are presented in the Article and are available from the corresponding authors upon reasonable request. REFERENCES * Du, E. et al. Global patterns of terrestrial nitrogen and phosphorus
limitation. _Nat. Geosci._ 13, 221–226 (2020). Article CAS ADS Google Scholar * Schulze, E. Ueber einige stickstoffhaltige Bestandtheile der Keimlinge von _Vicia sativa_. _Z. Phys.
Chem._ 17, 193–216 (1893). Google Scholar * Wishart, D. S. et al. HMDB 4.0: the human metabolome database for 2018. _Nucleic Acids Res._ 46, D608–D617 (2018). Article CAS PubMed Google
Scholar * Kato, T., Yamagata, M. & Tsukahara, S. Guanidine compounds in fruit trees and their seasonal variations in citrus (_Citrus unshiu_ Marc.). _J. Jpn. Soc. Hortic. Sci._ 55,
169–173 (1986). Article CAS Google Scholar * Gund, P. Guanidine, trimethylenemethane, and "Y-delocalization." Can acyclic compounds have "aromatic" stability? _J.
Chem. Educ._ 49, 100 (1972). Article CAS Google Scholar * Güthner, T., Mertschenk, B. & Schulz, B. In _Ullmann’s Fine Chemicals_ vol. 2, 657–672 (Wiley-VCH, 2014). * Strecker, A.
Untersuchungen über die chemischen Beziehungen zwischen Guanin, Xanthin, Theobromin, Caffeïn und Kreatinin. _Justus Liebigs Ann. Chem._ 118, 151–177 (1861). Article Google Scholar *
Iwanoff, N. N. & Awetissowa, A. N. The fermentative conversion of guanidine in urea. _Biochem. Z._ 231, 67–78 (1931). Google Scholar * Lenkeit, F., Eckert, I., Hartig, J. S. &
Weinberg, Z. Discovery and characterization of a fourth class of guanidine riboswitches. _Nucleic Acids Res._ 48, 12889–12899 (2020). Article CAS PubMed PubMed Central Google Scholar *
Salvail, H., Balaji, A., Yu, D., Roth, A. & Breaker, R. R. Biochemical validation of a fourth guanidine riboswitch class in bacteria. _Biochemistry_ 59, 4654–4662 (2020). Article CAS
PubMed Google Scholar * Nelson, J. W., Atilho, R. M., Sherlock, M. E., Stockbridge, R. B. & Breaker, R. R. Metabolism of free guanidine in bacteria is regulated by a widespread
riboswitch class. _Mol. Cell_ 65, 220–230 (2017). Article CAS PubMed Google Scholar * Sherlock, M. E. & Breaker, R. R. Biochemical validation of a third guanidine riboswitch class in
bacteria. _Biochemistry_ 56, 359–363 (2016). Article Google Scholar * Sherlock, M. E., Malkowski, S. N. & Breaker, R. R. Biochemical validation of a second guanidine riboswitch class
in bacteria. _Biochemistry_ 56, 352–358 (2016). Article Google Scholar * Kermani, A. A., Macdonald, C. B., Gundepudi, R. & Stockbridge, R. B. Guanidinium export is the primal function
of SMR family transporters. _Proc. Natl Acad. Sci. USA_ 115, 3060–3065 (2018). Article CAS PubMed PubMed Central Google Scholar * Sinn, M., Hauth, F., Lenkeit, F., Weinberg, Z. &
Hartig, J. S. Widespread bacterial utilization of guanidine as nitrogen source. _Mol. Microbiol._ 116, 200–210 (2021). Article CAS PubMed Google Scholar * Schneider, N. O. et al. Solving
the conundrum: widespread proteins annotated for urea metabolism in bacteria are carboxyguanidine deiminases mediating nitrogen assimilation from guanidine. _Biochemistry_ 59, 3258–3270
(2020). Article CAS PubMed Google Scholar * Zhao, J., Zhu, L., Fan, C., Wu, Y. & Xiang, S. Structure and function of urea amidolyase. _Biosci. Rep._ 38, BSR20171617 (2018). Article
CAS PubMed PubMed Central Google Scholar * Mobley, H. L., Island, M. D. & Hausinger, R. P. Molecular biology of microbial ureases. _Microbiol. Rev._ 59, 451–480 (1995). Article CAS
PubMed PubMed Central Google Scholar * Mazzei, L., Musiani, F. & Ciurli, S. The structure-based reaction mechanism of urease, a nickel dependent enzyme: tale of a long debate. _J.
Biol. Inorg. Chem._ 25, 829–845 (2020). Article CAS PubMed PubMed Central Google Scholar * Uribe, E. et al. Functional analysis of the Mn2+ requirement in the catalysis of
ureohydrolases arginase and agmatinase - a historical perspective. _J. Inorg. Biochem._ 202, 110812 (2020). Article CAS PubMed Google Scholar * Perozich, J., Hempel, J. & Morris, S.
M. Jr Roles of conserved residues in the arginase family. _Biochim. Biophys. Acta_ 1382, 23–37 (1998). Article CAS PubMed Google Scholar * Sekowska, A., Danchin, A. & Risler, J. L.
Phylogeny of related functions: the case of polyamine biosynthetic enzymes. _Microbiology_ 146, 1815–1828 (2000). Article CAS PubMed Google Scholar * Sekula, B. The neighboring subunit
is engaged to stabilize the substrate in the active site of plant arginases. _Front. Plant Sci._ 11, 987 (2020). Article PubMed PubMed Central Google Scholar * Quintero, M. J.,
Muro-Pastor, A. M., Herrero, A. & Flores, E. Arginine catabolism in the cyanobacterium _Synechocystis_ sp. strain PCC 6803 involves the urea cycle and arginase pathway. _J. Bacteriol._
182, 1008–1015 (2000). Article CAS PubMed PubMed Central Google Scholar * Lacasse, M. J., Summers, K. L., Khorasani-Motlagh, M., George, G. N. & Zamble, D. B. Bimodal nickel-binding
site on _Escherichia coli_ [NiFe]-hydrogenase metallochaperone HypA. _Inorg. Chem._ 58, 13604–13618 (2019). Article CAS PubMed PubMed Central Google Scholar * Hoffmann, D., Gutekunst,
K., Klissenbauer, M., Schulz-Friedrich, R. & Appel, J. Mutagenesis of hydrogenase accessory genes of _Synechocystis_ sp. PCC 6803. _FEBS J._ 273, 4516–4527 (2006). Article CAS PubMed
Google Scholar * Dowling, D. P., Di Costanzo, L., Gennadios, H. A. & Christianson, D. W. Evolution of the arginase fold and functional diversity. _Cell. Mol. Life Sci._ 65, 2039–2055
(2008). Article CAS PubMed PubMed Central Google Scholar * Dutta, A., Mazumder, M., Alam, M., Gourinath, S. & Sau, A. K. Metal-induced change in catalytic loop positioning in
_Helicobacter pylori_ arginase alters catalytic function. _Biochem. J._ 476, 3595–3614 (2019). Article CAS PubMed Google Scholar * Di Costanzo, L. et al. Crystal structure of human
arginase I at 1.29-Å resolution and exploration of inhibition in the immune response. _Proc. Natl Acad. Sci. USA_ 102, 13058–13063 (2005). Article PubMed PubMed Central ADS Google
Scholar * Suzek, B. E. et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. _Bioinformatics_ 31, 926–932 (2014). Article PubMed
PubMed Central Google Scholar * Alfano, M. & Cavazza, C. Structure, function, and biosynthesis of nickel-dependent enzymes. _Protein Sci._ 29, 1071–1089 (2020). Article CAS PubMed
PubMed Central Google Scholar * Wang, B. et al. A guanidine-degrading enzyme controls genomic stability of ethylene-producing cyanobacteria. _Nat. Commun._ 12, 5150 (2021). Article PubMed
PubMed Central ADS Google Scholar * McGee, D. J. et al. Purification and characterization of _Helicobacter pylori_ arginase, RocF: unique features among the arginase superfamily. _Eur.
J. Biochem._ 271, 1952–1962 (2004). Article CAS PubMed Google Scholar * Arakawa, N., Igarashi, M., Kazuoka, T., Oikawa, T. & Soda, K. d-Arginase of _Arthrobacter_ sp. KUJ 8602:
characterization and its identity with Zn2+-guanidinobutyrase. _J. Biochem._ 133, 33–42 (2003). Article CAS PubMed Google Scholar * Saragadam, T., Kumar, S. & Punekar, N. S.
Characterization of 4-guanidinobutyrase from _Aspergillus niger_. _Microbiology_ 165, 396–410 (2019). Article CAS PubMed Google Scholar * Viator, R. J., Rest, R. F., Hildebrandt, E.
& McGee, D. J. Characterization of _Bacillus anthracis_ arginase: effects of pH, temperature, and cell viability on metal preference. _BMC Biochem._ 9, 15 (2008). Article PubMed PubMed
Central Google Scholar * D’Antonio, E. L., Hai, Y. & Christianson, D. W. Structure and function of non-native metal clusters in human arginase I. _Biochemistry_ 51, 8399–8409 (2012).
Article PubMed Google Scholar * Andresen, E., Peiter, E. & Küpper, H. Trace metal metabolism in plants. _J. Exp. Bot._ 69, 909–954 (2018). Article CAS PubMed Google Scholar *
Eisenhut, M. Manganese homeostasis in cyanobacteria. _Plants_ 9, 18 (2019). Article PubMed Central Google Scholar * Burnat, M. & Flores, E. Inactivation of agmatinase expressed in
vegetative cells alters arginine catabolism and prevents diazotrophic growth in the heterocyst-forming cyanobacterium Anabaena. _MicrobiologyOpen_ 3, 777–792 (2014). Article CAS PubMed
PubMed Central Google Scholar * Callahan, B. P., Yuan, Y. & Wolfenden, R. The burden borne by urease. _J. Am. Chem. Soc._ 127, 10828–10829 (2005). Article CAS PubMed Google Scholar
* Lewis, C. A. Jr & Wolfenden, R. The nonenzymatic decomposition of guanidines and amidines. _J. Am. Chem. Soc._ 136, 130–136 (2014). Article CAS PubMed Google Scholar * Grobben,
Y. et al. Structural insights into human Arginase-1 pH dependence and its inhibition by the small molecule inhibitor CB-1158. _J. Struct. Biol. X_ 4, 100014 (2020). CAS PubMed Google
Scholar * Mills, L. A., McCormick, A. J. & Lea-Smith, D. J. Current knowledge and recent advances in understanding metabolism of the model cyanobacterium _Synechocystis_ sp. PCC 6803.
_Biosci. Rep._ 40, BSR20193325 (2020). Article CAS PubMed PubMed Central Google Scholar * Giner-Lamia, J. et al. Identification of the direct regulon of NtcA during early acclimation to
nitrogen starvation in the cyanobacterium _Synechocystis_ sp PCC 6803. _Nucleic Acids Res._ 45, 11800–11820 (2017). Article CAS PubMed PubMed Central Google Scholar * Martinez, S.
& Hausinger, R. P. Biochemical and spectroscopic characterization of the non-heme Fe(II)- and 2-oxoglutarate-dependent ethylene-forming enzyme from _Pseudomonas syringae_ pv.
_phaseolicola_ PK2. _Biochemistry_ 55, 5989–5999 (2016). Article CAS PubMed Google Scholar * Copeland, R. A. et al. An iron(IV)-oxo intermediate initiating l-arginine oxidation but not
ethylene production by the 2-oxoglutarate-dependent oxygenase, ethylene-forming enzyme. _J. Am. Chem. Soc._ 143, 2293–2303 (2021). Article CAS PubMed PubMed Central Google Scholar *
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M. & Stanier, R. Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. _Microbiology_ 111, 1–61
(1979). Article Google Scholar * Geyer, J. W. & Dabich, D. Rapid method for determination of arginase activity in tissue homogenates. _Anal. Biochem._ 39, 412–417 (1971). Article CAS
PubMed Google Scholar * van Anken, H. C. & Schiphorst, M. E. A kinetic determination of ammonia in plasma. _Clin. Chim. Acta_ 56, 151–157 (1974). Article PubMed Google Scholar *
Kabsch, W. XDS. _Acta Crystallogr. D_ 66, 125–132 (2010). Article CAS PubMed PubMed Central Google Scholar * McCoy, A. J. et al. Phaser crystallographic software. _J. Appl.
Crystallogr._ 40, 658–674 (2007). Article CAS PubMed PubMed Central Google Scholar * Lamzin, V. S. P. A., Wilson, K. S. In International Tables for Crystallography Vol. F (eds Arnold,
E. et al.) 525–528 (Kluwer, 2012). * Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. _Acta Crystallogr. D_ 66, 486–501 (2010). Article CAS PubMed
PubMed Central Google Scholar * Adams, P. D. et al. The Phenix software for automated determination of macromolecular structures. _Methods_ 55, 94–106 (2011). Article CAS PubMed PubMed
Central Google Scholar * Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. _Protein Sci._ 27, 293–315 (2018). Article CAS
Google Scholar * Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. _J. Mol. Graph. Model._ 25, 247–260
(2006). Article PubMed ADS Google Scholar * Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. _J. Chem. Theory Comput._ 11,
3696–3713 (2015). Article CAS PubMed PubMed Central Google Scholar * Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber
force field. _J. Comput. Chem._ 25, 1157–1174 (2004). Article CAS PubMed Google Scholar * Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a
new scoring function, efficient optimization, and multithreading. _J. Comput. Chem._ 31, 455–461 (2010). CAS PubMed PubMed Central Google Scholar * Ashkenazy, H. et al. ConSurf 2016: an
improved methodology to estimate and visualize evolutionary conservation in macromolecules. _Nucleic Acids Res._ 44, W344–W350 (2016). Article CAS PubMed PubMed Central Google Scholar *
Lemoine, F. et al. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. _Nature_ 556, 452–456 (2018). Article CAS PubMed PubMed Central ADS Google Scholar * Lemoine,
F. et al. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. _Nucleic Acids Res._ 47, W260–W265 (2019). Article CAS PubMed PubMed Central Google Scholar *
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. _Nucleic Acids Res._ 47, W256–W259 (2019). Article CAS PubMed PubMed Central Google
Scholar * Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. _Genome Res._ 14, 1188–1190 (2004). Article CAS PubMed PubMed Central Google
Scholar Download references ACKNOWLEDGEMENTS J.S.H. acknowledges the ERC CoG project 681777 “RiboDisc” for financial support. R.L.-I. was supported by grant PID2019-104784RJ-100
MCIN/AEI/10.13039/501100011033 Spain, and grant BFU2017-88202-P MCIN/AEI/10.13039/501100011033 Spain, cofinanced by ERDF A way of making Europe. We thank R. Winter and E. Flores for helpful
discussions and A. Joachimi and D. Galetskiy for technical assistance. We acknowledge the Paul Scherrer Institut, Villigen, Switzerland, for provision of synchrotron radiation beamtime at
beamline X06SA-PXI of the Swiss Light Source, and thank K. M. L. Smith for assistance. We thank K. Forchhammer for providing the _Synechocystis_ PCC 6803 GT cells and for helpful advice
regarding their cultivation. AUTHOR INFORMATION Author notes * These authors contributed equally: D. Funck, M. Sinn AUTHORS AND AFFILIATIONS * Department of Chemistry, University of
Konstanz, Konstanz, Germany D. Funck, M. Sinn, M. Stanoppi, J. Dietrich & J. S. Hartig * Department of Biology, University of Konstanz, Konstanz, Germany J. R. Fleming & O. Mayans *
Instituto de Bioquímica Vegetal y Fotosíntesis, Universidad de Sevilla and C.S.I.C, Seville, Spain R. López-Igual * Konstanz Graduate School Chemical Biology (KoRS-CB), University of
Konstanz, Konstanz, Germany O. Mayans & J. S. Hartig Authors * D. Funck View author publications You can also search for this author inPubMed Google Scholar * M. Sinn View author
publications You can also search for this author inPubMed Google Scholar * J. R. Fleming View author publications You can also search for this author inPubMed Google Scholar * M. Stanoppi
View author publications You can also search for this author inPubMed Google Scholar * J. Dietrich View author publications You can also search for this author inPubMed Google Scholar * R.
López-Igual View author publications You can also search for this author inPubMed Google Scholar * O. Mayans View author publications You can also search for this author inPubMed Google
Scholar * J. S. Hartig View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS D.F., M. Sinn and J.S.H. conceived the project. D.F. and M. Sinn
performed protein expression, purification, activity assays and sequence analyses. R.L.-I. generated the Δ_sll1077_ mutant and J.D., R.L.-I. and D.F. performed growth assays. M. Stanoppi
performed NMR analyses. J.R.F. and O.M. performed protein crystallization, structure determination, analysis and modelling. D.F., M. Sinn and J.S.H. wrote the manuscript with input from all
authors. D.F., M. Sinn, J.D., M. Stanoppi and J.R.F. prepared figures. The manuscript was reviewed and approved by all coauthors. CORRESPONDING AUTHOR Correspondence to J. S. Hartig. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature_ thanks David Richardson and the other, anonymous, reviewer(s) for
their contribution to the peer review of this work. Peer reviewer reports are available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional
claims in published maps and institutional affiliations. EXTENDED DATA FIGURES AND TABLES EXTENDED DATA FIG. 1 ADDITIONAL CHARACTERIZATION GDMH. A: Activation of GdmH expressed in the
absence of added metal ions and either co-expressed with GhaA and GhaB or with either of the accessory proteins alone by Ni2+ or Fe2+. The orange columns represent the activity in a mixture
of the latter two extracts. Where indicated, 1 mM β-mercaptoethanol (β-MSH) was additionally included. Columns represent the average of n = 3 technical replicates, error bars indicate s.d.
The entire experiment was repeated with independent bacterial extracts, which yielded slightly different absolute values for specific activities but virtually identical results for the
relative values. B: 13C-NMR spectra of 13C15N-guanidine after incubation over night with purified GdmH (overexpressed together with GhaA and GhaB). Exclusively the triplet signal of
13C15N-urea was detected after incubation with purified GdmH, whereas after incubation with GdmH partially inactivated by heat treatment, both the quadruplet of 13C15N-guanidine and the
triplet signal of 13C15N-urea were detected. C: Coupled enzymatic assay of GdmH with glutamate dehydrogenase (GDH). NADH gets oxidized by GDH during reductive amination of α-ketoglutarate
with ammonia released from guanidine by GdmH. Michaelis-constant _K__M_, maximal specific activity _A__max_, and catalytic constant _k__cat_ were determined using different guanidine
concentrations. Data points represent the average of n = 3 technical replicates and the black line represents the least-square fit to the Michaelis-Menten equation. Error bars represent s.d.
The whole experiment was repeated with an independent enzyme preparation, which gave consistent results. D: Influence of Na-cacodylate on the activity of GdmH with 10 mM guanidine as
substrate. A cacodylate molecule occupied the active site in one of the crystal structures but even a tenfold excess of cacodylate had no influence on GdmH activity. Columns represent the
average of n = 3 independent enzyme preparations, error bars indicate s.d. E: Effect of point mutations on the activity and _K__M_ of purified, recombinant GdmH. The black lines represent
the least square fits to the Michaelis-Menten equation of single experiments. The entire analysis was repeated with independent enzyme preparations with consistent results. The inset shows a
Coomassie-stained gel with the purified GdmH variants and the positions of molecular weight markers. For gel source data, see Supplementary Fig. 1b. F: Time-dependence of urea production by
GdmH in the presence of 10 mM guanidine. Note that almost half of the substrate was hydrolyzed after 3 days of incubation. EXTENDED DATA FIG. 2 ADDITIONAL IMAGES OF THE GDMH STRUCTURE. A:
Top view of the GdmH hexamer with one subunit in surface display (bright yellow) and two subunits as ribbons below a transparent surface (orange and sky blue). The extended N-terminus of the
orange subunit is highlighted by saturated color. B: Comparison of the GdmH capping helix (blue) with the same helix from the most similar protein with a high resolution structure,
guanidinobutyrase from _Pseudomonas aeruginosa_ (grey with capping helix in yellow, PDB entry 3NIO). Although the overall fold is well conserved between both enzymes, the position of the
highlighted α-helix is shifted towards the active site. EXTENDED DATA FIG. 3 RELATION OF GDMH TO OTHER PROTEINS FROM THE ARGINASE SUPERFAMILY. Unrooted neighbor-joining tree of 509
representatives of >15000 UniRef90 sequences with at least 27% identity to residues 60–390 of GdmH, selected and aligned with the ConSurf webserver61. Accession numbers of all sequences
are given in Supplementary Data 2. The branches are colored according to the taxonomic group: Black: bacteria; cyan: archaea; bright green: fungi; green: plants; brown: stramenopile;
magenta: animals. Branches containing selected enzymes with biochemically confirmed function are highlighted and demonstrate that very dissimilar sequences can have the same enzymatic
activity in different taxa. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIG. 1 Source data for SDS gel (Fig. 1e); source data for SDS gel (Extended Data Fig. 1e) as pdf file. REPORTING SUMMARY
PEER REVIEW FILE SUPPLEMENTARY DATA 1 Multiple-sequence alignment of GdmH with similar sequences as fasta formatted plain text file. SUPPLEMENTARY DATA 2 Manually optimized sequence
alignment of GdmH with further sequences of characterized enzymes of the arginase family (source data for Extended Data Fig. 3) as fasta formatted plain text file. SUPPLEMENTARY DATA 3
Multiple-sequence alignment of the 194 closest homologues of GdmH (source data for Fig. 3g) as fasta formatted plain text file. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS
ARTICLE CITE THIS ARTICLE Funck, D., Sinn, M., Fleming, J.R. _et al._ Discovery of a Ni2+-dependent guanidine hydrolase in bacteria. _Nature_ 603, 515–521 (2022).
https://doi.org/10.1038/s41586-022-04490-x Download citation * Received: 09 June 2021 * Accepted: 31 January 2022 * Published: 09 March 2022 * Issue Date: 17 March 2022 * DOI:
https://doi.org/10.1038/s41586-022-04490-x SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative