Recombinant elastomeric protein biopolymers: progress and prospects
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Genetically engineered protein biopolymers belong to a new family of polymers that have recently attracted interest due to their highly modifiable material properties. It is now
possible to use a bottom-up engineering process to design advanced, smart materials for biomedical and engineering applications, such as energy storage and bioremediation. This review
explores recent developments in these genetically engineered protein biopolymers, with a particular emphasis on elastomeric biopolymers. Also discussed are the future directions that this
field will likely explore. SIMILAR CONTENT BEING VIEWED BY OTHERS BIOINSPIRED AND BIOMIMETIC PROTEIN-BASED FIBERS AND THEIR APPLICATIONS Article Open access 18 April 2024 RESILIN-MIMETICS AS
A SMART BIOMATERIAL PLATFORM FOR BIOMEDICAL APPLICATIONS Article Open access 08 January 2021 DESIGN AND SUSTAINABILITY OF POLYPEPTIDE MATERIAL SYSTEMS Article 14 April 2025 INTRODUCTION
Genetically engineered protein biopolymers have recently attracted tremendous attention for various applications in engineering and materials science. With advances in recombinant
technologies, it is now possible to have precise control over the genetic sequence and properties of a given protein biopolymer. As such, recombinant protein biopolymers can be specially
designed to possess multifunctionalities, including self-assembling abilities, directed biological activities and even unique metal binding properties. These characteristics make recombinant
proteins particularly useful for the preparation of a wide variety of functional materials for regenerative medicine and engineering applications. In this focused review, we aim to provide
an overview of the current prospects in elastomeric protein biopolymer research. ELASTIN Elastin is one of the most abundant extracellular matrix proteins found in organs where elasticity is
of major importance, such as large arteries, elastic ligaments, lungs and skin.1, 2, 3, 4 Elastin-like polypeptides (ELPs) consist of repetitive peptide sequences derived from the
hydrophobic domain of mammalian tropoelastin, the precursor protein of elastin. The most commonly studied motif of ELPs is the pentapeptide motif (VPGXG)_m_, where X is known as a guest
amino acid other than proline. The subscript _m_ describes the number of repeats, which typically ranges between 20 and 330 repeats.5 Following pioneering studies on the development and
characterization of ELPs by Urry and group,6, 7 elastin-based biopolymers have been heavily investigated for a wide variety of applications. A unique property of ELPs is their ability to
undergo a sharp and reversible phase transition at a specific temperature known as the inverse transition temperature (_T_t) or the lower critical solution temperature (LCST); both of these
terms can be used interchangeably.8 ELPs undertake hydrophilic random coil conformations below their LCST and are highly soluble in water. Above their LCST, ELPs aggregate rapidly into
micron-size particles that are visible to the naked eye.9 Such phase transitions are reversible and can be rapidly triggered by temperature shifts. Urry and group6, 7 demonstrated that this
LCST is highly dependent on the amino acid composition of the peptide repeat. Therefore, the LCST can be tailored to suit the application by changing the guest residue X in the pentapeptide
motif (VPGXG)_m_.5 Moreover, this inverse transition temperature enables ELPs to be easily purified simply by repeated centrifugation while cycling the temperature above and below its LCST;
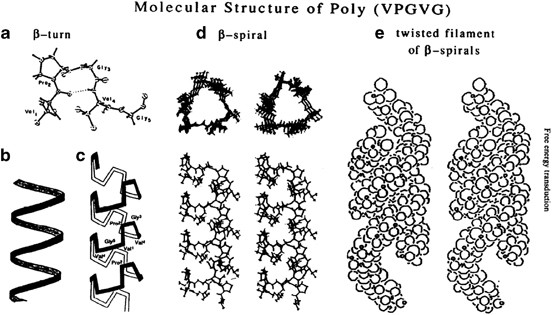
this technique is termed ITC or inverse thermal cycling.10 Upon changes in temperature, pH and salt concentration, ELPs self-assemble into organized network structures11 (Figure 1). Such
self-assembling behavior is particularly useful for tissue engineering and drug delivery applications. ELPs can also serve as templates for nanomaterial synthesis.12, 13, 14 ELPS IN TISSUE
ENGINEERING Tissue engineering has evolved into a multidisciplinary field involving materials science, engineering and biology. To serve as functional substitutes for damaged tissues,
biomaterials are now expected to mimic the biological, mechanical and topographical characteristics of the native tissues they are replacing. ELP biopolymers are attractive candidates for
tissue engineering for several reasons. First, ELPs are known to have excellent biocompatibility and are degradable into natural amino acids that cause a minimum amount of cytotoxicity,
immune response and inflammation.10, 15, 16, 17 One of the earliest examples of ELP use in tissue engineering was work performed by Setton and Chilkoti.18 In this case, ELPs were used to
create an injectable three-dimensional matrix to entrap chondrocytes for cartilaginous tissue repair.18 _In vitro_ studies revealed that the encapsulated chondrocytes retained a rounded
morphology and a chondrocytic phenotype. The authors demonstrated that when human adipose-derived stem cells were encapsulated, ELP gels induce chondrocytic differentiation, even in the
absence of chondrocyte-specific growth factors.18 A similar study was also performed by Haider _et al._,19 where human mesenchymal stem cells were used for chondrogenesis. These reports
suggest that ELP-based biopolymers have great potential for use in cartilage tissue repair. However, uncrosslinked ELPs exhibit shear modulus several orders lower than cartilage. Therefore,
their mechanical integrity is insufficient to match and support functional cartilage repair. ELPs have also been chemically crosslinked to produce scaffolds with improved mechanical
integrity using well-established crosslinking methods. Common examples include the following: tissue transglutaminase, which catalyzes covalent bond formation between glutamine residues and
primary amines; the amine reactive crosslinker β-[tris(hydroxymethyl) phosphino] propionic acid; and an NHS-ester crosslinker that crosslinks primary amines (of lysine and arginine
residues).20, 21, 22 The use of tissue transglutaminase to crosslink ELP is favorable; gelation occurs under physiological conditions, and the reaction can be performed _in situ_ with little
effect on living encapsulated cells. However, this crosslinking reaction proceeded over a long period of time, limiting its clinical use. Similarly, Nagapudi _et al._23 also reported the
use of photoactive methacrylate polymers for the crosslinking of ELPs under UV and visible light for cell encapsulation. In addition to cell encapsulation, other uses of ELPs in tissue
engineering include tissue-engineered vascular grafts. Tirrell and group24 designed a small diameter vascular graft using ELPs fused with fibronectin CS5 domains. The CS5 domain present
within the fusion construct enhanced endothelial cell adhesion.25, 26 Crosslinked films made from the CS5-containing ELPs also exhibited mechanical properties similar to that of native
elastin (elastic moduli ranges between 0.3 and 0.6 MPa).17 ELP-based biopolymers can be designed to have increased and more integrin-specific bioactivities through the incorporation of
selective cell-binding domains. For example, ELPs containing the full-length fibronectin domains 9 and 10 were shown to engage alpha5beta1 integrins with increased affinities, leading to
faster wound closure.27 Recently, Annabi _et al._28 reported the use of an elastin-based biopolymer in cardiac tissue engineering. Specifically, a methacrylated tropoelastin hydrogel was
developed that had high resilience (approximately 400%) upon stretching and deformed reversibly with minimum energy loss.29 In this work, the micropatterned methacrylated tropoelastin
hydrogel was also shown to direct the alignment of cardiomyocytes and synchronize their contractile properties _in vitro_28 (Figure 2), mimicking those of native myocardium. Although elastin
is found abundantly in skin, only several reports on the application of elastin-based biomaterials as skin substitutes have been completed. One such work by Rnjak-Kovacina _et al._30 used
electrospun recombinant human tropoelastin mixed with bovine collagen to create composite porous scaffolds for use as dermal substitutes. Their _in vivo_ cell culture results demonstrated
that the blending of both proteins supported cell infiltration, cell proliferation and new capillary formation when compared with Integra, a commercially available dermal substitute.
Nonetheless, most of the literature is focused on designing the optimal scaffold topography or architecture for dermal fibroblasts. Our group has designed and produced ELP fusion biopolymers
specifically for artificial skin substrates. In our work, each ELP fusion protein sequence was designed to contain cell-binding domains derived from native fibronectin, laminin and
collagen. Such fusion constructs were designed to target major integrin–extracellular matrix interactions utilized by human skin keratinocytes. We found that keratinocyte interactions with
the various cell-binding domains present in each ELP resulted in differing rates of keratinocytes adhesion and motility.31 Hence, it is possible to design cell-instructive biomaterials that
also mimic the microenvironment of human tissues. ELPS FOR THE SYNTHESIS OF INORGANIC MATERIALS The concept of using of ELP fusion proteins for metal binding was first demonstrated by Chen
and coworkers.32, 33, 34, 35 Specifically, ELP fusion proteins capable of recognizing and binding heavy metals were developed for the purpose of bioremediation. In their reports, ELP fusion
proteins were able to bind heavy metals in water and were subsequently recovered by utilizing the temperature-responsive characteristics of ELPs.32 Chen and workers33, 34 also demonstrated
the efficiency of ELPs containing two metal-binding domains, polyhistidine and phytochelatin, in cadmium removal. When phytochelatin was used as the metal-binding domain for _ex situ_ soil
washing, a fivefold increase in cadmium removal was observed compared to polyhistidine. Phytochelatin exhibited higher binding affinity and binding capacity compared to cadmium. In another
study, 90% of cadmium was removed in less than an hour using a low concentration of 0.25 mM ELP biopolymer; this was compared to 4 mM EDTA.33, 34 Subsequent studies on other metal-binding
domains have also been reported, including a study on the bacterial metalloregulatory protein MerR, which has a very high affinity to mercury. In this work, it was reported that the
retrieval of mercury in Lake Elsinore, CA, USA, water was highly selective even in the presence of unrelated heavy metals at an excess of 100-fold.35 ELPs were also explored as templates for
the synthesis of inorganic metallic nanoparticles.36, 37 For example, ELPs containing silver-binding domains were designed and used for the biomimetic synthesis of silver nanoparticles.
Specifically, the fusion protein GPG–AG3 discovered by Naik and group,39 which contains a tropoelastin-like domain (that is, GPG38) and the AG3 sequence (AYSSGAPPMPPF), possessed the ability
to reduce choloaurate ([AuCl4]3−) to nucleate gold nanoparticles or Ag+ to Ag0 under physiological conditions.40 In a report by Anh _et al._,40 GPG–AG3 fusion proteins presented in the form
of protein aggregates or crosslinked spun-coated thin films were able to nucleate silver nanoparticles from soluble silver precursors (Figure 3). Both forms of GPG–AG3 materials containing
nucleated silver nanoparticles possess antibacterial properties,40 demonstrating promising use as antibacterial wound dressings and biomedical implant coatings. Apart from their biomedical
applications, ELPs also hold promise for use in the biomineralization of inorganic nanomaterials. Biomineralization is a natural phenomenon that occurs in living organisms. It involves a
combination of concerted mechanisms including bioaccumulation, reduction and mineralization processes to form hard tissues or to protect them from metal toxicity.41, 42 There has been
increasing advancement in the creation of multifunctional hierarchical inorganic composites using biological systems as templates, which have been inspired by native biomineralization
processes. For example, wire-shaped flagella from bacteria and rod-like viruses (for example, the tobacco mosaic virus and M13) have been used or modified with metal-binding motifs to
biomimetically fabricate various metal nanowire/tube structures.43 In our laboratory, we have explored the use of ELPs as a versatile template for the fabrication of long-range noble metal
nanostructures. For example, Guo _et al._44 have successfully utilized the self-assembling GPG–AG3 fusion protein for the biomineralization of noble metal nanoparticles. In this work, the
porous nanofibers Pt and Pd were obtained and had excellent catalytic capabilities (manuscript under review). While ELPs and ELP fusion proteins are a promising class of biomaterials for
tissue engineering and regenerative medicine, the use of ELPs for other important engineering applications such as bioremediation, energy storage and catalysis is also being explored. SILK
Silk is another class of materials with unique mechanical properties. Silk and silk-like biopolymers have high strength-to-weight ratios and mechanical properties that span a wide range of
moduli (3–10 GPa).45 Silk proteins contain highly repetitive protein sequences that can be crystalline or non-crystalline in nature. For example, the crystalline region from silkworm _Bombyx
mori_ fibroin is mostly composed of glycine, alanine and serine in a 3:2:1 ratio (that is, GAGAGSGAAG[SGAGAG]8Y).46 Likewise, the repetitive motifs of spider silk spindroin from orb-weaving
_Nephila clavipes_ dragline silk (YGGLGSQGAGRGG) have also been identified.47, 48 The versatile mechanical properties of silk make silk-like biopolymers very attractive for use in
biomedical applications. _B. mori_ silk fibers, for example, have been used as sutures since the end of 19th century. Dragline spider silk has also been heavily studied for its high
strength-to-weight ratio. However, the translation of silk-based biomaterials to clinical use has been limited by difficulties in harvesting native silk proteins. It is particularly
difficult to obtain scalable amounts of dragline silk because the large-scale farming of spiders is not possible. For this reason, recombinant silk proteins and silk-like biopolymers have
gained popularity; large amounts of materials can be biosynthetically produced in various host systems, including bacteria,49 insect cells,50, 51 mammalian cells52 and yeast.53 Many
synthetic genes encoding _B. mori_ silk-like sequences, such as the six amino acid repeat GAGAGS, [(AG)3PEG] and [(AG)3EG], have been cloned and expressed in bacterial hosts such as
_Escherichia coli_.49, 54, 55 Engineered spindroins have also been developed and recombinantly produced in _E. coli_; these include the ADF3 and ADF4 sequences from _Araneus diadematus_.56,
57 Recombinant spindroins have also been produced in large quantities, with protein yields up to 360 mg l−1, using bacterial fermentation methods.58 Therefore, the use of such proteins on an
industrial scale is promising. Like ELPs, the increasing popularity of silk has stimulated a widespread interest in silk-based fusion proteins. For example, fusion proteins containing silk
and extracellular matrix motifs derived from collagen, laminin and fibronectin have been developed. Such proteins have been shown to increase the attachment and spreading of various cells
compared to those cultured on unmodified silk nanofibers.59, 60, 61, 62, 63 In terms of materials processing, it has also been shown that recombinant spindroins have the capability to be
processed into a wide range of morphologies (Figure 4).64, 65 To date, silk-based biopolymers have been investigated for use as biomaterial scaffolds for the following: skin66, 67; skeletal
tissue such as cartilage,62, 68, 69, 70 bone71, 72 and ligaments/tendons73; vascular tissues74; and nerve grafts.75 With continuing advancement in recombinant technologies, silk-based
biopolymers will likely continue to be an interesting and important class of materials in tissue engineering. RESILIN Resilin is another structural protein that is recently of interest in
tissue engineering research, largely due to its unique mechanical properties. Resilin is an elastomeric protein found commonly in the ligament and tendons of insects.76 In a previous study,
resilin isolated from locust tendons was able to store large amounts of energy with little loss to heat.77 Native resilin is composed of random coils, crosslinked by di- and tri-tyrosine
links. This crosslinking results in a stable and flexible three-dimensional network, conferring to resilin its elastomeric properties.78 The elasticity of resilin is largely attributed to
the high content of glycine and proline residues, which form β-turn and polyproline II conformations.79, 80 Like most structural proteins previously discussed, a recombinant resilin-like
biopolymer could also be reproduced using _E. coli_ expression hosts. The _CG15920_ gene from _Drosophila melanogaster_ is one of the most popular domains found in recombinant resilin-like
biopolymers.81 Particularly, the repetitive motif GGRPSDSYGAPGGGN located in the N-terminal region of CG15920 has been widely studied. For instance, Kaplan and group82 examined the protein
exons 1 and 3; exon 1 exhibited 90% resilience compared to 63%, which made exon 1 the chosen domain to mimic native resilin. The authors also sought an alternative di-tyrosine formation
technique for resilin-based hydrogels using a citrate-modified photo-Fenton system. The study revealed interesting adhesive properties for the crosslinked hydrogels, as
L-3,4-dihydroxyphenylalanie was also formed. This crosslinked resilin-based hydrogel displayed a high resilience of 90% by withstanding over 50 cycles of extension using an atomic force
microscope82 (Figure 5). Li and Kiick80 demonstrated that recombinant resilin-like hydrogels have comparable mechanical strength and extensibility to native forms. The authors added RGD
cell-binding domains to the resilin-like polypeptides (RLPs), and crosslinked the fusion proteins using Tris(hydroxymethyl phosphine). The authors were able to obtain mechanically stable
hydrogels, which could maintain the viability of primary human mesenchymal stem cells and mouse fibroblast NIH-3T3 cells.83, 84 Separately, composite hydrogels consisting of resilin-like
polypeptides and polyethylene glycol were developed by designing cysteine residues in resilin-like polypeptides as crosslinking sites for the vinyl sulfone groups in polyethylene glycol;
these hydrogels were able to support the viability and spreading of encapsulated human aortic adventitial fibroblasts.85 Likewise, hybrid biopolymers containing resilin-like, elastin-like
and collagen-like domains were also reported to have bioactivity.86 Recently, Li and group87, 88, 89 reported on an artificial protein containing periodic repeats of GB1 and resilin (Figure
6). The GB1 polyprotein is derived from the streptococcal B1 immunoglobulin-binding domain of protein G. This polypeptide has been shown to possess mechanical stability over long periods of
continuous stretching-relaxation cycles.87 In their work, crosslinked GB1–resilin scaffolds had an elastic modulus of approximately 50 kPa (at 15% strain) and could be stretched up to
135%.89 In a follow-up study, these authors demonstrated that the elastic stiffness of crosslinked GB1–resilin scaffolds could also be fine-tuned by controlling the crosslinking density.
This yielded scaffolds with moduli ranging between 10 and 65 kPa.90 TITIN Titin is a major component of human muscle sarcomeres; it is also the largest single polypeptide (approximately 4
MDa) known in nature.91, 92, 93, 94, 95, 96 Native titin consists of long linear chains of approximately 300 modular domains, of which 90% are structured. Of these structured domains, the
most widely studied are the fibronectin-like type III and the immunoglobulin-like domains. In contrast, the unstructured domains are flexible and rich in proline (P), glutamate (E), valine
(V) and lysine (K) (and referred to as PEVK).92 Native titin has been widely studied to better understand the mechanisms responsible for the extensibility and the elastic behavior of human
muscle. Single-molecule force studies have shown that the high mechanical strength, fracture toughness and the elasticity of titin are due to the sequential folding/unfolding of its
structured domains at the molecular level.97, 98, 99, 100, 101, 102 It has been suggested that the unfolding of certain immunoglobulin-like and PEVK domains is responsible for the passive
elasticity observed in muscle.92, 94, 103, 104 Specifically, the immunoglobulin-like 27 domain in titin is composed of 89 amino acids and adopts a β-sandwich fold structure105; this domain
can be unfolded with a characteristic force of 204 pN.97 It was further proposed that the mechanical force to resist unfolding in immunoglobulin-like 27 is provided by hydrogen bonds between
two parallel terminal β-strands in a sandwich formation.105 Conversely, the randomly folded PEVK domain contains 75% proline, glutamate, valine and lysine residues. It has been proposed
that the unfolding of PEVK domains accounts for the flexible and extensible behavior of human muscle.94, 106, 107 Given the unique extensibility of titin, titin-based biopolymers have also
been investigated. Li _et al._97 have produced a series of recombinant proteins containing various immunoglobulin-like and PEVK domains and studied their mechanical properties using
single-molecule force microscopy. In this work, the authors found that the immunoglobulin-like 27 domain unfolded at a force of 204 pN. Conversely, the extension of PEVK and N2B (unique
sequences of tandem immunoglobulin-like and PEVK segments) regions in cardiac I-band titin was observed to be an average of 68 and 209 nm, respectively. The collective extension contributed
by these random domains measured by single-molecule force microscopy is 300 nm, consistent with cardiac myofibril data97 (Figure 7). These findings suggest that it may be possible to
recreate recombinant biopolymers that have muscle-like mechanical properties. At present, research on titin-based biopolymers is largely focused on understanding their mechanical behavior at
the single-molecule level. However, it is likely that scaffolds derived from titin-based biopolymers have tremendous potential in tissue-engineering applications, given their unique
muscle-like properties. SUMMARY The ability to tailor both the chemical and physical properties of recombinant biopolymers has created a wide range of opportunities in the end applications
of these materials. This review aims to provide the scientific community with an overview of current trends in the field of recombinantly engineered elastomeric protein biopolymers. In this
article, we summarized current research on some biopolymers, and we described ongoing work on unpublished recombinant biopolymers developed in our laboratory. We also evaluated their
potential applications in biomedicine and other engineering disciplines, such as inorganic nanomaterial synthesis and bioremediation. REFERENCES * Wise, S. G., Mithieux, S. M., Raftery, M.
J. & Weiss, A. S. Specificity in the coacervation of tropoelastin: solvent exposed lysines. _J. Struct. Biol._ 149, 273–281 (2005). Article CAS PubMed Google Scholar * Vrhovski, B.,
Jensen, S. & Weiss, A. S. Coacervation characteristics of recombinant human tropoelastin. _Eur. J. Biochem._ 250, 92–98 (1997). Article CAS PubMed Google Scholar * Vrhovski, B. &
Weiss, A. S. Biochemistry of tropoelastin. _Eur. J. Biochem._ 258, 1–18 (1998). Article CAS PubMed Google Scholar * Bellingham, C. M., Lillie, M. A., Gosline, J. M., Wright, G. M.,
Starcher, B. C., Bailey, A. J., Woodhouse, K. A. & Keeley, F. W. Recombinant human elastin polypeptides self-assemble into biomaterials with elastin-like properties. _Biopolymers_ 70,
445–455 (2003). Article CAS PubMed Google Scholar * Meyer, D. E. & Chilkoti, A. Quantification of the effects of chain length and concentration on the thermal behavior of
elastin-like polypeptides. _Biomacromolecules_ 5, 846–851 (2004). Article CAS PubMed Google Scholar * Urry, D. W. & Long, M. M. Conformations of the repeat peptides of elastin in
solution: an application of proton and carbon-13 magnetic resonance to the determination of polypeptide secondary structure. _Crit. Rev. Biochem._ 4, 1–45 (1976). Article CAS Google
Scholar * McPherson, D. T., Morrow, C., Minehan, D. S., Wu, J., Hunter, E. & Urry, D. W. Production and purification of a recombinant elastomeric polypeptide, G-(VPGVG)19-VPGV, from
_Escherichia coli_. _Biotechnol. Prog._ 8, 347–352 (1992). Article CAS PubMed Google Scholar * Urry, D. W. Physical chemistry of biological free energy transduction as demonstrated by
elastic protein-based polymers. _J. Phys. Chem. B_ 101, 11007–11028 (1997). Article CAS Google Scholar * Meyer, D. E. & Chilkoti, A. Genetically encoded synthesis of protein-based
polymers with precisely specified molecular weight and sequence by recursive directional ligation: examples from the elastin-like polypeptide system. _Biomacromolecules_ 3, 357–367 (2002).
Article CAS PubMed Google Scholar * Meyer, D. E., Trabbic, C. K. & Chilkoti, A. Protein purification by fusion with an environmentally responsive elastin-like polypeptide: effect of
polypeptide length on the purification of thioredoxin. _Biotechnol. Prog._ 17, 720–728 (2001). Article CAS PubMed Google Scholar * Urry, D. W. Free energy transduction in polypeptides
and proteins based on inverse temperature transitions. _Prog. Biophys. Mol. Biol._ 57, 23–57 (1992). Article CAS PubMed Google Scholar * Wright, E. R., McMillan, R. A., Cooper, A.,
Apkarian, R. P. & Conticello, V. P. Thermoplastic elastomer hydrogels via self-assembly of an elastin-mimetic triblock polypeptide. _Adv. Funct. Mater._ 12, 149–154 (2002). Article CAS
Google Scholar * MacKay, J. A., Chen, M., McDaniel, J. R., Liu, W., Simnick, A. J. & Chilkoti, A. Self-assembling chimeric polypeptide-doxorubicin conjugate nanoparticles that abolish
tumours after a single injection. _Nat. Mater._ 8, 993–999 (2009). Article PubMed PubMed Central CAS Google Scholar * Zhang, S. Fabrication of novel biomaterials through molecular
self-assembly. _Nat. Biotechnol._ 21, 1171–1178 (2003). Article CAS PubMed Google Scholar * Megeed, Z., Haider, M., Li, D., O'Malley, B. W. Jr, Cappello, J. & Ghandehari, H. _In
vitro_ and _in vivo_ evaluation of recombinant silk-elastinlike hydrogels for cancer gene therapy. _J. Control. Rel._ 94, 433–445 (2004). Article CAS Google Scholar * Urry, D. W. Elastic
molecular machines in metabolism and soft-tissue restoration. _Trends Biotechnol._ 17, 249–257 (1999). Article CAS PubMed Google Scholar * Nicol, A., Gowda, D. C., Parker, T. M. &
Urry, D. W. Elastomeric polytetrapeptide matrices: hydrophobicity dependence of cell attachment from adhesive (GGIP)n to nonadhesive (GGAP)n even in serum. _J. Biomed. Mater. Res._ 27,
801–810 (1993). Article CAS PubMed Google Scholar * Betre, H., Ong, S. R., Guilak, F., Chilkoti, A., Fermor, B. & Setton, L. A. Chondrocytic differentiation of human adipose-derived
adult stem cells in elastin-like polypeptide. _Biomaterials_ 27, 91–99 (2006). Article CAS PubMed Google Scholar * Haider, M., Cappello, J., Ghandehari, H. & Leong, K. W. _In vitro_
chondrogenesis of mesenchymal stem cells in recombinant silk-elastinlike hydrogels. _Pharm. Res._ 25, 692–699 (2008). Article CAS PubMed Google Scholar * McHale, M. K., Setton, L. A.
& Chilkoti, A. Synthesis and _in vitro_ evaluation of enzymatically cross-linked elastin-like polypeptide gels for cartilaginous tissue repair. _Tissue Eng._ 11, 1768–1779 (2005).
Article CAS PubMed Google Scholar * Nettles, D. L., Haider, M. A., Chilkoti, A. & Setton, L. A. Neural network analysis identifies scaffold properties necessary for _in vitro_
chondrogenesis in elastin-like polypeptide biopolymer scaffolds. _Tissue Eng. Part A_ 16, 11–20 (2010). Article CAS PubMed Google Scholar * Lim, D. W., Nettles, D. L., Setton, L. A.
& Chilkoti, A. _In situ_ cross-linking of elastin-like polypeptide block copolymers for tissue repair. _Biomacromolecules_ 9, 222–230 (2008). Article CAS PubMed Google Scholar *
Nagapudi, K., Brinkman, W. T., Leisen, J. E., Huang, L., McMillan, R. A., Apkarian, R. P., Conticello, V. P. & Chaikof, E. L. Photomediated solid-state cross-linking of an
elastin−mimetic recombinant protein polymer. _Macromolecules_ 35, 1730–1737 (2002). Article CAS Google Scholar * Heilshorn, S. C., DiZio, K. A., Welsh, E. R. & Tirrell, D. A.
Endothelial cell adhesion to the fibronectin CS5 domain in artificial extracellular matrix proteins. _Biomaterials_ 24, 4245–4252 (2003). Article CAS PubMed Google Scholar * Liu, J. C.,
Heilshorn, S. C. & Tirrell, D. A. Comparative cell response to artificial extracellular matrix proteins containing the RGD and CS5 cell-binding domains. _Biomacromolecules_ 5, 497–504
(2004). Article CAS PubMed Google Scholar * Welsh, E. R. & Tirrell, D. A. Engineering the extracellular matrix: a novel approach to polymeric biomaterials. I. Control of the physical
properties of artificial protein matrices designed to support adhesion of vascular endothelial cells. _Biomacromolecules_ 1, 23–30 (2000). Article CAS PubMed Google Scholar * Fong, E.
& Tirrell, D. A. Collective cell migration on artificial extracellular matrix proteins containing full-length fibronectin domains. _Adv. Mater._ 22, 5271–5275 (2010). Article CAS
PubMed PubMed Central Google Scholar * Annabi, N., Tsang, K., Mithieux, S. M., Nikkhah, M., Ameri, A., Khademhosseini, A. & Weiss, A. S. Highly elastic micropatterned hydrogel for
engineering functional cardiac tissue. _Adv. Funct. Mater._ 23, 4950–4959 (2013). Article CAS Google Scholar * Annabi, N., Mithieux, S. M., Zorlutuna, P., Camci-Unal, G., Weiss, A. S.
& Khademhosseini, A. Engineered cell-laden human protein-based elastomer. _Biomaterials_ 34, 5496–5505 (2013). Article CAS PubMed PubMed Central Google Scholar * Rnjak-Kovacina, J.,
Wise, S. G., Li, Z., Maitz, P. K. M., Young, C. J., Wang, Y. & Weiss, A. S. Electrospun synthetic human elastin:collagen composite scaffolds for dermal tissue engineering. _Acta
Biomaterialia_ 8, 3714–3722 (2012). Article CAS PubMed Google Scholar * Tjin, M. S., Chua, A. W. C., Ma, D. R., Lee, S. T. & Fong, E. Human epidermal keratinocyte cell response on
integrin-specific artificial extracellular matrix proteins. _Macromol. Biosci._ (e-pub ahead of print 2 May 2014; doi:10.1002/mabi.201400015). * Kostal, J., Prabhukumar, G., Lao, U. L.,
Chen, A., Matsumoto, M., Mulchandani, A. & Chen, W. Customizable biopolymers for heavy metal remediation. _J. Nanopart. Res._ 7, 517–523 (2005). Article CAS Google Scholar * Lao, U.
L., Chen, A., Matsumoto, M. R., Mulchandani, A. & Chen, W. Cadmium removal from contaminated soil by thermally responsive elastin (ELPEC20) biopolymers. _Biotechnol. Bioeng._ 98, 349–355
(2007). Article CAS PubMed Google Scholar * Prabhukumar, G., Matsumoto, M., Mulchandani, A. & Chen, W. Cadmium removal from contaminated soil by tunable biopolymers. _Environ. Sci.
Technol._ 38, 3148–3152 (2004). Article CAS PubMed Google Scholar * Kostal, J., Mulchandani, A., Gropp, K. E. & Chen, W. A temperature responsive biopolymer for mercury remediation.
_Environ. Sci. Technol._ 37, 4457–4462 (2003). Article CAS PubMed Google Scholar * Naik, R. R., Stringer, S. J., Agarwal, G., Jones, S. E. & Stone, M. O. Biomimetic synthesis and
patterning of silver nanoparticles. _Nat. Mater._ 1, 169–172 (2002). Article CAS PubMed Google Scholar * Kim, J., Sadowsky, M. J. & Hur, H. G. Simultaneous synthesis of
temperature-tunable peptide and gold nanoparticle hybrid spheres. _Biomacromolecules_ 12, 2518–2523 (2011). Article CAS PubMed Google Scholar * Le, D. H. T., Hanamura, R., Pham, D.-H.,
Kato, M., Tirrell, D. A., Okubo, T. & Sugawara-Narutaki, A. Self-assembly of elastin–mimetic double hydrophobic polypeptides. _Biomacromolecules_ 14, 1028–1034 (2013). Article CAS
PubMed Google Scholar * Slocik, J. M., Stone, M. O. & Naik, R. R. Synthesis of gold nanoparticles using multifunctional peptides. _Small_ 1, 1048–1052 (2005). Article CAS PubMed
Google Scholar * Anh, T. T., Xing, M., Le, D. H., Sugawara-Narutaki, A. & Fong, E. Elastin-based silver-binding proteins with antibacterial capabilities. _Nanomedicine (Lond)_ 8,
567–575 (2013). Article CAS Google Scholar * Addadi, L. & Weiner, S. Control and design principles in biological mineralization. _Angew. Chem. Int. Ed._ 31, 153–169 (1992). Article
Google Scholar * Addadi, L. & Weiner, S. Crystals, asymmetry and life. _Nature_ 411, 753–755 (2001). Article CAS PubMed Google Scholar * Rong, J., Niu, Z., Lee, L. A. & Wang, Q.
Self-assembly of viral particles. _Curr. Opin. Colloid Interface Sci._ 16, 441–450 (2011). Article CAS Google Scholar * Guo, G., Truong, T. H.A., Tan, H., Ang, H., Zhang, W., Xu, C.,
Rui, X., Hu, Z., Fong, E. & Yan, Q. Platinum and Palladium Nanotubes Based on Genetically Engineered Elastin–Mimetic Fusion Protein-Fiber Templates: Synthesis and Application in
Lithium-O2 Batteries, Chemistry. _An Asian Journal_ (in press). * Gosline, J., Lillie, M., Carrington, E., Guerette, P., Ortlepp, C. & Savage, K. Elastic proteins: biological roles and
mechanical properties. _Phil. Trans. R. Soc. Lond. B_ 357, 121–132 (2002). Article CAS Google Scholar * Tsujimoto, Y. & Suzuki, Y. Structural analysis of the fibroin gene at the
5' end and its surrounding regions. _Cell_ 16, 425–436 (1979). Article CAS PubMed Google Scholar * Mello, C. M., Senecal, K., Yeung, B., Vouros, P. & Kaplan, D. (eds) Initial
characterization of _Nephila clavipes_ dragline protein in _Silk Polymers_ VOL. 544, 67–79 (American Chemical Society, Washington, DC, 1994). Google Scholar * Xu, M. & Lewis, R. V.
Structure of a protein superfiber: spider dragline silk. _Proc. Natl. Acad. Sci._ 87, 7120–7124 (1990). Article CAS PubMed Google Scholar * Cappello, J., Crissman, J., Dorman, M.,
Mikolajczak, M., Textor, G., Marquet, M. & Ferrari, F. Genetic engineering of structural protein polymers. _Biotechnol. Prog._ 6, 198–202 (1990). Article CAS PubMed Google Scholar *
Wen, H., Lan, X., Zhang, Y., Zhao, T., Wang, Y., Kajiura, Z. & Nakagaki, M. Transgenic silkworms (_Bombyx mori_) produce recombinant spider dragline silk in cocoons. _Mol. Biol. Rep._
37, 1815–1821 (2010). Article CAS PubMed Google Scholar * Zhang, Y., Hu, J., Miao, Y., Zhao, A., Zhao, T., Wu, D., Liang, L., Miikura, A., Shiomi, K., Kajiura, Z. & Nakagaki, M.
Expression of EGFP-spider dragline silk fusion protein in BmN cells and larvae of silkworm showed the solubility is primary limit for dragline proteins yield. _Mol. Biol. Rep._ 35, 329–335
(2008). Article PubMed CAS Google Scholar * Lazaris, A., Arcidiacono, S., Huang, Y., Zhou, J.-F., Duguay, F., Chretien, N., Welsh, E. A., Soares, J. W. & Karatzas, C. N. Spider silk
fibers spun from soluble recombinant silk produced in mammalian cells. _Science_ 298, 472–476 (2002). Article Google Scholar * Fahnestock, S. R. & Bedzyk, L. A. Production of synthetic
spider dragline silk protein in _Pichia pastoris_. _Appl. Microbiol. Biotechnol._ 47, 33–39 (1997). Article CAS PubMed Google Scholar * Krejchi, M. T., Atkins, E., Waddon, A. J.,
Fournier, M. J., Mason, T. L. & Tirrell, D. A. Chemical sequence control of beta-sheet assembly in macromolecular crystals of periodic polypeptides. _science_ 265, 1427–1432 (1994).
Article CAS PubMed Google Scholar * McGrath, K. P., Fournier, M. J., Mason, T. L. & Tirrell, D. A. Genetically directed syntheses of new polymeric materials. Expression of artificial
genes encoding proteins with repeating-(AlaGly)3ProGluGly- elements. _J. Am. Chem. Soc._ 114, 727–733 (1992). Article CAS Google Scholar * Huemmerich, D., Helsen, C. W., Quedzuweit, S.,
Oschmann, J., Rudolph, R. & Scheibel, T. Primary structure elements of spider dragline silks and their contribution to protein solubility. _Biochemistry_ 43, 13604–13612 (2004). Article
CAS PubMed Google Scholar * Rammensee, S., Slotta, U., Scheibel, T. & Bausch, A. R. Assembly mechanism of recombinant spider silk proteins. _Proc. Natl Acad. Sci._ 105, 6590–6595
(2008). Article CAS PubMed Google Scholar * Scheibel, T. Spider silks: recombinant synthesis, assembly, spinning, and engineering of synthetic proteins. _Microb. Cell Fact._ 3, 14
(2004). Article PubMed PubMed Central CAS Google Scholar * Min, B.-M., Jeong, L., Nam, Y. S., Kim, J.-M., Kim, J. Y. & Park, W. H. Formation of silk fibroin matrices with different
texture and its cellular response to normal human keratinocytes. _International J. Biol. Macromol._ 34, 223–230 (2004). Article CAS Google Scholar * Huang, F., Sun, L. & Zheng, J. _In
vitro_ and _in vivo_ characterization of a silk fibroin-coated polyester vascular prosthesis. _Art. Organs_ 32, 932–941 (2008). Article Google Scholar * Fan, H., Liu, H., Wong, E. J. W.,
Toh, S. L. & Goh, J. C. H. _In vivo_ study of anterior cruciate ligament regeneration using mesenchymal stem cells and silk scaffold. _Biomaterials_ 29, 3324–3337 (2008). Article CAS
PubMed Google Scholar * Meinel, L., Hofmann, S., Karageorgiou, V., Zichner, L., Langer, R., Kaplan, D. & Vunjak-Novakovic, G. Engineering cartilage-like tissue using human mesenchymal
stem cells and silk protein scaffolds. _Biotechnol. Bioeng._ 88, 379–391 (2004). Article CAS PubMed Google Scholar * Meinel, L., Karageorgiou, V., Hofmann, S., Fajardo, R., Snyder, B.,
Li, C., Zichner, L., Langer, R., Vunjak-Novakovic, G. & Kaplan, D. L. Engineering bone-like tissue _in vitro_ using human bone marrow stem cells and silk scaffolds. _J. Biomed. Mater.
Res. A_ 71A, 25–34 (2004). Article CAS Google Scholar * Humenik, M., Smith, A. M. & Scheibel, T. Recombinant spider silk—biopolymers with potential for future applications. _Polymers_
3, 640–661 (2011). Article CAS Google Scholar * Vepari, C. & Kaplan, D. L. Silk as a biomaterial. _Prog. Polym. Sci._ 32, 991–1007 (2007). Article CAS PubMed PubMed Central
Google Scholar * Sugihara, A., Sugiura, K., Morita, H., Ninagawa, T., Tubouchi, K., Tobe, R., Izumiya, M., Horio, T., Abraham, N. G. & Ikehara, S. Promotive effects of a silk film on
epidermal recovery from full-thickness skin wounds. _Proc. Soc. Exp. Biol. Med._ 225, 58–64 (2000). Article CAS PubMed Google Scholar * Roh, D.-H., Kang, S.-Y., Kim, J.-Y., Kwon, Y.-B.,
Young Kweon, H., Lee, K.-G., Park, Y.-H., Baek, R.-M., Heo, C.-Y., Choe, J. & Lee, J.-H. Wound healing effect of silk fibroin/alginate-blended sponge in full thickness skin defect of
rat. _J. Mater. Sci. Mater. Med._ 17, 547–552 (2006). Article CAS PubMed Google Scholar * Wang, Y., Blasioli, D. J., Kim, H.-J., Kim, H. S. & Kaplan, D. L. Cartilage tissue
engineering with silk scaffolds and human articular chondrocytes. _Biomaterials_ 27, 4434–4442 (2006). Article CAS PubMed Google Scholar * Wang, Y., Kim, U.-J., Blasioli, D. J., Kim,
H.-J. & Kaplan, D. L. _In vitro_ cartilage tissue engineering with 3D porous aqueous-derived silk scaffolds and mesenchymal stem cells. _Biomaterials_ 26, 7082–7094 (2005). Article CAS
PubMed Google Scholar * Marolt, D., Augst, A., Freed, L. E., Vepari, C., Fajardo, R., Patel, N., Gray, M., Farley, M., Kaplan, D. & Vunjak-Novakovic, G. Bone and cartilage tissue
constructs grown using human bone marrow stromal cells, silk scaffolds and rotating bioreactors. _Biomaterials_ 27, 6138–6149 (2006). Article CAS PubMed Google Scholar * Sofia, S.,
McCarthy, M. B., Gronowicz, G. & Kaplan, D. L. Functionalized silk-based biomaterials for bone formation. _J. Biomed. Mater. Res._ 54, 139–148 (2001). Article CAS PubMed Google
Scholar * Wang, X., Kim, H. J., Xu, P., Matsumoto, A. & Kaplan, D. L. Biomaterial coatings by stepwise deposition of silk fibroin. _Langmuir_ 21, 11335–11341 (2005). Article CAS
PubMed Google Scholar * Altman, G. H., Horan, R. L., Lu, H. H., Moreau, J., Martin, I., Richmond, J. C. & Kaplan, D. L. Silk matrix for tissue engineered anterior cruciate ligaments.
_Biomaterials_ 23, 4131–4141 (2002). Article CAS PubMed Google Scholar * Ma, X., Cao, C. & Zhu, H. The biocompatibility of silk fibroin films containing sulfonated silk fibroin. _J.
Biomed. Mater. Res. B_ 78B, 89–96 (2006). Article CAS Google Scholar * Yang, Y., Chen, X., Ding, F., Zhang, P., Liu, J. & Gu, X. Biocompatibility evaluation of silk fibroin with
peripheral nerve tissues and cells _in vitro_. _Biomaterials_ . 28, 1643–1652 (2007). Article PubMed CAS Google Scholar * Weis-Fogh, T. A rubber-like protein in insect cuticle. _J. Exp.
Biol._ 37, 889–907 (1960). CAS Google Scholar * Su, R. S. C., Kim, Y. & Liu, J. C. Resilin: Protein-based elastomeric biomaterials. _Acta Biomaterialia_ 10, 1601–1611 (2013). Article
PubMed CAS Google Scholar * Kappiyoor, R., Balasubramanian, G., Dudek, D. M. & Puri, I. K. Elastomechanical properties of resilin. _Soft Matter_ 7, 11006–11009 (2011). Article CAS
Google Scholar * Renner, J. N., Kim, Y., Cherry, K. M. & Liu, J. C. Modular cloning and protein expression of long, repetitive resilin-based proteins. _Protein Expr. Purif._ 82, 90–96
(2012). Article CAS PubMed Google Scholar * Li, L. & Kiick, K. L. Resilin-based materials for biomedical applications. _ACS Macro Lett._ 2, 635–640 (2013). Article CAS PubMed
PubMed Central Google Scholar * Li, L., Charati, M. B. & Kiick, K. L. Elastomeric polypeptide-based biomaterials. _Polym. Chem._ 1, 1160–1170 (2010). Article CAS Google Scholar *
Qin, G., Rivkin, A., Lapidot, S., Hu, X., Preis, I., Arinus, S. B., Dgany, O., Shoseyov, O. & Kaplan, D. L. Recombinant exon-encoded resilins for elastomeric biomaterials. _Biomaterials_
32, 9231–9243 (2011). Article CAS PubMed PubMed Central Google Scholar * Li, L., Tong, Z., Jia, X. & Kiick, K. L. Resilin-like polypeptide hydrogels engineered for versatile
biological function. _Soft Matter_ 9, 665–673 (2013). Article CAS PubMed Google Scholar * Li, L., Teller, S., Clifton, R. J., Jia, X. & Kiick, K. L. Tunable mechanical stability and
deformation response of a resilin-based elastomer. _Biomacromolecules_ 12, 2302–2310 (2011). Article CAS PubMed PubMed Central Google Scholar * McGann, C. L., Levenson, E. A. &
Kiick, K. L. Resilin-based hybrid hydrogels for cardiovascular tissue engineering. _Macromol. Chem. Phys._ 214, 203–213 (2013). Article CAS Google Scholar * Bracalello, A., Santopietro,
V., Vassalli, M., Marletta, G., Del Gaudio, R., Bochicchio, B. & Pepe, A. Design and production of a chimeric resilin-, elastin-, and collagen-like engineered polypeptide.
_Biomacromolecules_ 12, 2957–2965 (2011). Article CAS PubMed Google Scholar * Cao, Y. & Li, H. B. Polyprotein of GB1 is an ideal artificial elastomeric protein. _Nat. Mater._ 6,
109–114 (2007). Article CAS PubMed Google Scholar * Elvin, C. M., Carr, A. G., Huson, M. G., Maxwell, J. M., Pearson, R. D., Vuocolo, T., Liyou, N. E., Wong, D. C., Merritt, D. J. &
Dixon, N. E. Synthesis and properties of crosslinked recombinant pro-resilin. _Nature_ 437, 999–1002 (2005). Article CAS PubMed Google Scholar * Lv, S., Dudek, D. M., Cao, Y.,
Balamurali, M. M., Gosline, J. & Li, H. B. Designed biomaterials to mimic the mechanical properties of muscles. _Nature_ 465, 69–73 (2010). Article CAS PubMed Google Scholar * Fang,
J. & Li, H. B. A facile way to tune mechanical properties of artificial elastomeric proteins-based hydrogels. _Langmuir_ 28, 8260–8265 (2012). Article CAS PubMed Google Scholar *
Furst, B. (ed). Functional morphology of the heart in _The Heart and Circulation_ 95–109 (Springer, London, 2014). Chapter Google Scholar * Granzier, H. L. & Labeit, S. The giant
protein titin—a major player in myocardial mechanics, signaling, and disease. _Circ. Res._ 94, 284–295 (2004). Article CAS PubMed Google Scholar * Linke, W. A., Rudy, D. E., Centner, T.,
Gautel, M., Witt, C., Labeit, S. & Gregorio, C. C. I-band titin in cardiac muscle is a three-element molecular spring and is critical for maintaining thin filament structure. _J. Cell
Biol._ 146, 631–644 (1999). Article CAS PubMed PubMed Central Google Scholar * Hsin, J., Strumpfer, J., Lee, E. H. & Schulten, K. Molecular origin of the hierarchical elasticity of
titin: simulation, experiment, and theory. _Ann. Rev. Biophys._ 40, 187–203 (2011). Article CAS Google Scholar * Radke, M. H., Peng, J., Wu, Y. M., McNabb, M., Nelson, O. L., Granzier, H.
& Gotthardt, M. Targeted deletion of titin N2B region leads to diastolic dysfunction and cardiac atrophy. _Proc. Natl. Acad. Sci. USA_ 104, 3444–3449 (2007). Article CAS PubMed
Google Scholar * Anderson, B. R. & Granzier, H. L. Titin-based tension in the cardiac sarcomere: molecular origin and physiological adaptations. _Progr. Biophys. Mol. Biol._ 110,
204–217 (2012). Article CAS Google Scholar * Li, H. B., Linke, W. A., Oberhauser, A. F., Carrion-Vazquez, M., Kerkvliet, J. G., Lu, H., Marszalek, P. E. & Fernandez, J. M. Reverse
engineering of the giant muscle protein titin. _Nature_ 418, 998–1002 (2002). Article CAS PubMed Google Scholar * Javadi, Y., Fernandez, J. M. & Perez-Jimenez, R. Protein folding
under mechanical forces: a physiological view. _Physiology_ 28, 9–17 (2013). Article CAS PubMed Google Scholar * Tskhovrebova, L., Trinick, J., Sleep, J. & Simmons, R. Elasticity and
unfolding of single molecules of the giant muscle protein titin. _Nature_ 387, 308–312 (1997). Article CAS PubMed Google Scholar * Labeit, S. & Kolmerer, B. Titins: giant proteins
in charge of muscle ultrastructure and elasticity. _Science_ 270, 293–296 (1995). Article CAS PubMed Google Scholar * Granzier, H., Helmes, M. & Trombitás, K. Nonuniform elasticity
of titin in cardiac myocytes: a study using immunoelectron microscopy and cellular mechanics. _Biophys. J._ 70, 430–442 (1996). Article CAS PubMed PubMed Central Google Scholar *
Erickson, H. P. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. _Proc. Natl Acad.
Sci. USA_ 91, 10114–10118 (1994). Article CAS PubMed Google Scholar * Carrion-Vazquez, M., Oberhauser, A. F., Fowler, S. B., Marszalek, P. E., Broedel, S. E., Clarke, J. & Fernandez,
J. M. Mechanical and chemical unfolding of a single protein: a comparison. _Proc. Natl Acad. Sci. USA_ 96, 3694–3699 (1999). Article CAS PubMed Google Scholar * Fukuda, N., Granzier, H.
L., Ishiwata, S. & Kurihara, S. Physiological functions of the giant elastic protein titin in mammalian striated muscle. _J. Phys. Sci._ 58, 151–159 (2008). Article CAS Google Scholar
* Duff, N., Duong, N. H. & Lacks, D. J. Stretching the immunoglobulin 27 domain of the titin protein: the dynamic energy landscape. _Biophys. J._ 91, 3446–3455 (2006). Article CAS
PubMed PubMed Central Google Scholar * Caamano, S., Sarkar, A. & Fernandez, J. M. The elasticity of individual titin PEVK exons measured by single molecule atomic force microscopy.
_Biophys. J._ 88, 169a–169a (2005). Google Scholar * Duan, Y., DeKeyser, J. G., Damodaran, S. & Greaser, M. L. Studies on titin PEVK peptides and their interaction (vol 454, pg 16,
2006). _Arch. Biochem. Biophys._ 456, 232–232 (2006). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We acknowledge funding from the Ministry of Education AcRF Tier 1
(RG41). Tjin and Low are supported by the Research Student Scholarship (RSS) from Nanyang Technological University, Singapore. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * School of
Materials Science and Engineering, Nanyang Technological University, Singapore, Singapore Monica S Tjin, Pearlie Low & Eileen Fong Authors * Monica S Tjin View author publications You
can also search for this author inPubMed Google Scholar * Pearlie Low View author publications You can also search for this author inPubMed Google Scholar * Eileen Fong View author
publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Eileen Fong. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS
ARTICLE CITE THIS ARTICLE Tjin, M., Low, P. & Fong, E. Recombinant elastomeric protein biopolymers: progress and prospects. _Polym J_ 46, 444–451 (2014).
https://doi.org/10.1038/pj.2014.65 Download citation * Received: 28 February 2014 * Revised: 23 May 2014 * Accepted: 23 May 2014 * Published: 02 July 2014 * Issue Date: August 2014 * DOI:
https://doi.org/10.1038/pj.2014.65 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative